Gene Page: FGF17
Summary ?
| GeneID | 8822 |
| Symbol | FGF17 |
| Synonyms | FGF-13|FGF-17|HH20 |
| Description | fibroblast growth factor 17 |
| Reference | MIM:603725|HGNC:HGNC:3673|Ensembl:ENSG00000158815|HPRD:04765|Vega:OTTHUMG00000097051 |
| Gene type | protein-coding |
| Map location | 8p21 |
| Pascal p-value | 0.709 |
| Fetal beta | -0.745 |
| eGene | Cerebellar Hemisphere Cerebellum |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.031 | |
| GSMA_IIA | Genome scan meta-analysis (All samples) | Psr: 0.03086 | |
| GSMA_IIE | Genome scan meta-analysis (European-ancestry samples) | Psr: 0.00057 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
General gene expression (GTEx)
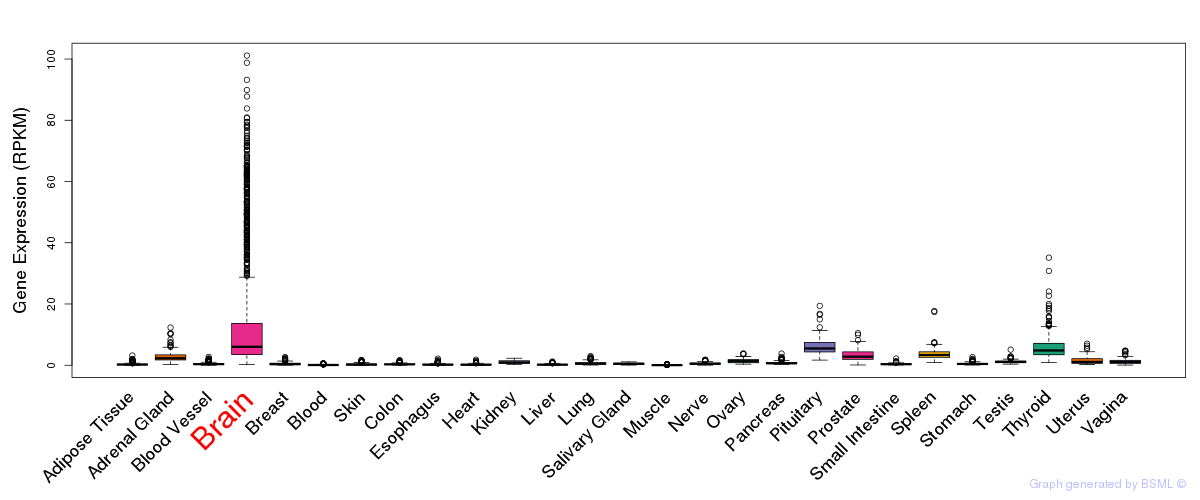
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0008083 | growth factor activity | TAS | 9514906 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007399 | nervous system development | TAS | neurite (GO term level: 5) | 9514906 |
| GO:0007267 | cell-cell signaling | TAS | 9514906 | |
| GO:0007165 | signal transduction | TAS | 9514906 | |
| GO:0008284 | positive regulation of cell proliferation | IEA | - | |
| GO:0008543 | fibroblast growth factor receptor signaling pathway | EXP | 17133345 | |
| GO:0008543 | fibroblast growth factor receptor signaling pathway | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005576 | extracellular region | EXP | 1697263 |9139660 |11294897 |16597617 | |
| GO:0005576 | extracellular region | NAS | 12975309 | |
| GO:0005615 | extracellular space | TAS | 9514906 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG MAPK SIGNALING PATHWAY | 267 | 205 | All SZGR 2.0 genes in this pathway |
| KEGG REGULATION OF ACTIN CYTOSKELETON | 216 | 144 | All SZGR 2.0 genes in this pathway |
| KEGG PATHWAYS IN CANCER | 328 | 259 | All SZGR 2.0 genes in this pathway |
| KEGG MELANOMA | 71 | 57 | All SZGR 2.0 genes in this pathway |
| PID FGF PATHWAY | 55 | 37 | All SZGR 2.0 genes in this pathway |
| REACTOME NEGATIVE REGULATION OF FGFR SIGNALING | 37 | 24 | All SZGR 2.0 genes in this pathway |
| REACTOME INSULIN RECEPTOR SIGNALLING CASCADE | 87 | 64 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY FGFR IN DISEASE | 127 | 88 | All SZGR 2.0 genes in this pathway |
| REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | 16 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY FGFR1 MUTANTS | 30 | 20 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | 11 | 8 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY FGFR3 MUTANTS | 11 | 9 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY FGFR MUTANTS | 44 | 29 | All SZGR 2.0 genes in this pathway |
| REACTOME FRS2 MEDIATED CASCADE | 36 | 27 | All SZGR 2.0 genes in this pathway |
| REACTOME PI 3K CASCADE | 56 | 39 | All SZGR 2.0 genes in this pathway |
| REACTOME DOWNSTREAM SIGNALING OF ACTIVATED FGFR | 100 | 74 | All SZGR 2.0 genes in this pathway |
| REACTOME PHOSPHOLIPASE C MEDIATED CASCADE | 54 | 40 | All SZGR 2.0 genes in this pathway |
| REACTOME SHC MEDIATED CASCADE | 28 | 19 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY INSULIN RECEPTOR | 108 | 72 | All SZGR 2.0 genes in this pathway |
| REACTOME FGFR LIGAND BINDING AND ACTIVATION | 22 | 15 | All SZGR 2.0 genes in this pathway |
| REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | 12 | 9 | All SZGR 2.0 genes in this pathway |
| REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | 12 | 8 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY FGFR | 112 | 80 | All SZGR 2.0 genes in this pathway |
| REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | 14 | 9 | All SZGR 2.0 genes in this pathway |
| REACTOME PI3K CASCADE | 71 | 51 | All SZGR 2.0 genes in this pathway |
| LEE NEURAL CREST STEM CELL DN | 118 | 79 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE FIMA UP | 544 | 308 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE LPS UP | 431 | 237 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3 UNMETHYLATED | 536 | 296 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K27ME3 | 269 | 159 | All SZGR 2.0 genes in this pathway |
| WONG ADULT TISSUE STEM MODULE | 721 | 492 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MEF HCP WITH H3K27ME3 | 590 | 403 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE UP | 857 | 456 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 2ND EGF PULSE ONLY | 1725 | 838 | All SZGR 2.0 genes in this pathway |
| NABA SECRETED FACTORS | 344 | 197 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME ASSOCIATED | 753 | 411 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME | 1028 | 559 | All SZGR 2.0 genes in this pathway |