Gene Page: CCKAR
Summary ?
| GeneID | 886 |
| Symbol | CCKAR |
| Synonyms | CCK-A|CCK1-R|CCK1R|CCKRA |
| Description | cholecystokinin A receptor |
| Reference | MIM:118444|HGNC:HGNC:1570|Ensembl:ENSG00000163394|HPRD:00322|Vega:OTTHUMG00000128567 |
| Gene type | protein-coding |
| Map location | 4p15.2 |
| Pascal p-value | 0.069 |
| Fetal beta | -0.105 |
| DMG | 1 (# studies) |
| eGene | Nucleus accumbens basal ganglia |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWAScat | Genome-wide Association Studies | This data set includes 560 SNPs associated with schizophrenia. A total of 486 genes were mapped to these SNPs within 50kb. | |
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 2 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Association | A combined odds ratio method (Sun et al. 2008), association studies | 2 | Link to SZGene |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 3 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg18636319 | 4 | 26492206 | CCKAR | 3.723E-4 | -0.444 | 0.043 | DMG:Wockner_2014 |
| cg08845028 | 4 | 26491963 | CCKAR | 5.114E-4 | -0.433 | 0.047 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
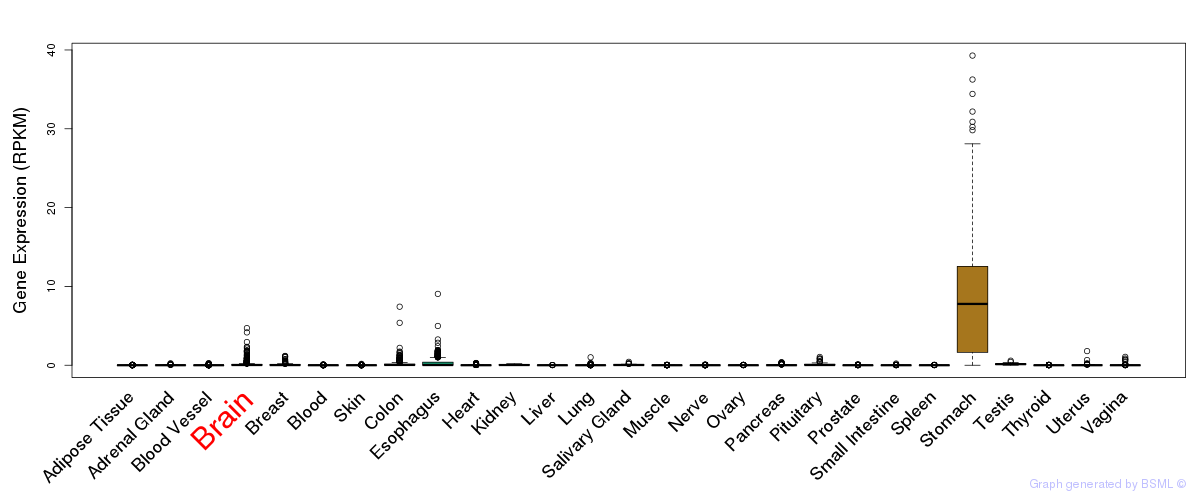
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| PRC1 | 0.96 | 0.75 |
| ZWINT | 0.96 | 0.78 |
| AURKA | 0.95 | 0.69 |
| NUSAP1 | 0.95 | 0.59 |
| KIF4A | 0.95 | 0.62 |
| BUB1 | 0.95 | 0.59 |
| CCNB2 | 0.95 | 0.53 |
| BIRC5 | 0.94 | 0.55 |
| RAD51 | 0.94 | 0.63 |
| KIF2C | 0.94 | 0.57 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.27 | -0.44 | -0.61 |
| AF347015.8 | -0.44 | -0.63 |
| AF347015.15 | -0.44 | -0.63 |
| MT-CYB | -0.44 | -0.61 |
| AF347015.31 | -0.44 | -0.59 |
| AF347015.33 | -0.44 | -0.60 |
| MT-CO2 | -0.43 | -0.59 |
| AF347015.2 | -0.43 | -0.62 |
| AF347015.26 | -0.42 | -0.62 |
| AF347015.9 | -0.40 | -0.55 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004872 | receptor activity | IEA | - | |
| GO:0004951 | cholecystokinin receptor activity | TAS | 8343165 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007409 | axonogenesis | IEA | neuron, axon, neurite (GO term level: 12) | - |
| GO:0001764 | neuron migration | IEA | neuron (GO term level: 8) | - |
| GO:0030900 | forebrain development | IEA | Brain (GO term level: 8) | - |
| GO:0007200 | G-protein signaling, coupled to IP3 second messenger (phospholipase C activating) | TAS | 8343165 | |
| GO:0007204 | elevation of cytosolic calcium ion concentration | TAS | 8503909 | |
| GO:0007165 | signal transduction | IEA | - | |
| GO:0007631 | feeding behavior | TAS | 8503909 | |
| GO:0007586 | digestion | TAS | 8503909 | |
| GO:0007584 | response to nutrient | TAS | 8503909 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005886 | plasma membrane | TAS | 8343165 | |
| GO:0005887 | integral to plasma membrane | TAS | 8343165 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CALCIUM SIGNALING PATHWAY | 178 | 134 | All SZGR 2.0 genes in this pathway |
| KEGG NEUROACTIVE LIGAND RECEPTOR INTERACTION | 272 | 195 | All SZGR 2.0 genes in this pathway |
| REACTOME GASTRIN CREB SIGNALLING PATHWAY VIA PKC AND MAPK | 205 | 136 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY GPCR | 920 | 449 | All SZGR 2.0 genes in this pathway |
| REACTOME PEPTIDE LIGAND BINDING RECEPTORS | 188 | 108 | All SZGR 2.0 genes in this pathway |
| REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | 305 | 177 | All SZGR 2.0 genes in this pathway |
| REACTOME G ALPHA Q SIGNALLING EVENTS | 184 | 116 | All SZGR 2.0 genes in this pathway |
| REACTOME GPCR DOWNSTREAM SIGNALING | 805 | 368 | All SZGR 2.0 genes in this pathway |
| REACTOME GPCR LIGAND BINDING | 408 | 246 | All SZGR 2.0 genes in this pathway |
| ODONNELL METASTASIS DN | 24 | 13 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 48HR DN | 428 | 306 | All SZGR 2.0 genes in this pathway |
| SHEN SMARCA2 TARGETS DN | 357 | 212 | All SZGR 2.0 genes in this pathway |
| SATO SILENCED BY METHYLATION IN PANCREATIC CANCER 1 | 419 | 273 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER RELAPSE IN BRAIN UP | 39 | 20 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS DN | 435 | 289 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH INV 16 TRANSLOCATION | 422 | 277 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS UP | 682 | 433 | All SZGR 2.0 genes in this pathway |
| RAO BOUND BY SALL4 ISOFORM A | 182 | 108 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 2ND EGF PULSE ONLY | 1725 | 838 | All SZGR 2.0 genes in this pathway |