Gene Page: PER3
Summary ?
| GeneID | 8863 |
| Symbol | PER3 |
| Synonyms | GIG13 |
| Description | period circadian clock 3 |
| Reference | MIM:603427|HGNC:HGNC:8847|Ensembl:ENSG00000049246|HPRD:09138|Vega:OTTHUMG00000001216 |
| Gene type | protein-coding |
| Map location | 1p36.23 |
| Pascal p-value | 0.238 |
| Sherlock p-value | 0.296 |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Association | A combined odds ratio method (Sun et al. 2008), association studies | 1 | Link to SZGene |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenics,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs16829545 | chr2 | 151977407 | PER3 | 8863 | 8.034E-5 | trans | ||
| rs4265582 | chr11 | 18797649 | PER3 | 8863 | 0.17 | trans | ||
| rs16955618 | chr15 | 29937543 | PER3 | 8863 | 6.618E-4 | trans |
Section II. Transcriptome annotation
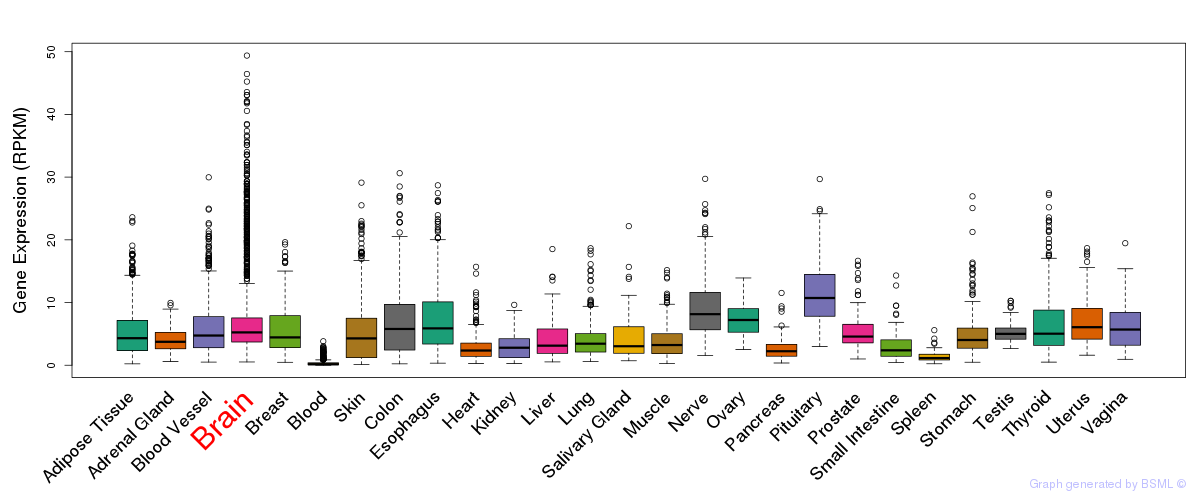
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| PPEF1 | 0.57 | 0.54 |
| THOC7 | 0.48 | 0.51 |
| TCEAL1 | 0.48 | 0.52 |
| FABP3 | 0.47 | 0.56 |
| TM6SF1 | 0.46 | 0.52 |
| GPR160 | 0.45 | 0.49 |
| MMD | 0.45 | 0.47 |
| UPP2 | 0.45 | 0.45 |
| C2orf80 | 0.44 | 0.46 |
| ACTR10 | 0.43 | 0.45 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| PLXNB1 | -0.32 | -0.39 |
| CASKIN2 | -0.31 | -0.39 |
| SORBS3 | -0.31 | -0.37 |
| PXN | -0.30 | -0.37 |
| ABTB2 | -0.29 | -0.36 |
| GPR56 | -0.29 | -0.37 |
| KIAA0195 | -0.29 | -0.31 |
| SH3BP4 | -0.29 | -0.34 |
| PLCB3 | -0.29 | -0.32 |
| LLGL1 | -0.28 | -0.30 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004871 | signal transducer activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006355 | regulation of transcription, DNA-dependent | IEA | - | |
| GO:0006350 | transcription | IEA | - | |
| GO:0007165 | signal transduction | IEA | - | |
| GO:0007623 | circadian rhythm | IEA | - | |
| GO:0048511 | rhythmic process | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005737 | cytoplasm | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CIRCADIAN RHYTHM MAMMAL | 13 | 13 | All SZGR 2.0 genes in this pathway |
| PYEON HPV POSITIVE TUMORS UP | 98 | 47 | All SZGR 2.0 genes in this pathway |
| LIU PROSTATE CANCER DN | 481 | 290 | All SZGR 2.0 genes in this pathway |
| THUM SYSTOLIC HEART FAILURE DN | 244 | 147 | All SZGR 2.0 genes in this pathway |
| HORIUCHI WTAP TARGETS UP | 306 | 188 | All SZGR 2.0 genes in this pathway |
| ODONNELL TFRC TARGETS UP | 456 | 228 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS DN | 459 | 276 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED DN | 805 | 505 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE EPIDERMIS DN | 508 | 354 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 2A DN | 141 | 84 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 4 5WK DN | 196 | 131 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 6 7WK DN | 79 | 54 | All SZGR 2.0 genes in this pathway |
| RIZ ERYTHROID DIFFERENTIATION | 77 | 51 | All SZGR 2.0 genes in this pathway |
| RIZ ERYTHROID DIFFERENTIATION CCNE1 | 40 | 26 | All SZGR 2.0 genes in this pathway |
| LASTOWSKA NEUROBLASTOMA COPY NUMBER DN | 800 | 473 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER APOCRINE VS LUMINAL | 326 | 213 | All SZGR 2.0 genes in this pathway |
| SHETH LIVER CANCER VS TXNIP LOSS PAM6 | 46 | 32 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS UP | 811 | 508 | All SZGR 2.0 genes in this pathway |
| CHESLER BRAIN QTL CIS | 75 | 51 | All SZGR 2.0 genes in this pathway |
| AFFAR YY1 TARGETS DN | 234 | 137 | All SZGR 2.0 genes in this pathway |
| LINDVALL IMMORTALIZED BY TERT DN | 80 | 56 | All SZGR 2.0 genes in this pathway |
| HENDRICKS SMARCA4 TARGETS DN | 53 | 34 | All SZGR 2.0 genes in this pathway |
| JIANG HYPOXIA CANCER | 83 | 52 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL CULTURED VS FRESH UP | 425 | 298 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS 60HR UP | 293 | 203 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA CHEMOTAXIS DN | 457 | 302 | All SZGR 2.0 genes in this pathway |
| PYEON CANCER HEAD AND NECK VS CERVICAL UP | 193 | 95 | All SZGR 2.0 genes in this pathway |
| CHANDRAN METASTASIS DN | 306 | 191 | All SZGR 2.0 genes in this pathway |
| OHGUCHI LIVER HNF4A TARGETS UP | 44 | 30 | All SZGR 2.0 genes in this pathway |
| GABRIELY MIR21 TARGETS | 289 | 187 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS UP | 504 | 321 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 DN | 918 | 550 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS UP | 745 | 475 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION DN | 1080 | 713 | All SZGR 2.0 genes in this pathway |
| SERVITJA LIVER HNF1A TARGETS UP | 135 | 96 | All SZGR 2.0 genes in this pathway |
| PLASARI TGFB1 TARGETS 10HR DN | 244 | 157 | All SZGR 2.0 genes in this pathway |
| ZWANG DOWN BY 2ND EGF PULSE | 293 | 119 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 1ST EGF PULSE ONLY | 1839 | 928 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-194 | 361 | 368 | 1A,m8 | hsa-miR-194 | UGUAACAGCAACUCCAUGUGGA |
| miR-29 | 1925 | 1931 | m8 | hsa-miR-29aSZ | UAGCACCAUCUGAAAUCGGUU |
| hsa-miR-29bSZ | UAGCACCAUUUGAAAUCAGUGUU | ||||
| hsa-miR-29cSZ | UAGCACCAUUUGAAAUCGGU | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.