Gene Page: SQSTM1
Summary ?
| GeneID | 8878 |
| Symbol | SQSTM1 |
| Synonyms | A170|FTDALS3|OSIL|PDB3|ZIP3|p60|p62|p62B |
| Description | sequestosome 1 |
| Reference | MIM:601530|HGNC:HGNC:11280|Ensembl:ENSG00000161011|HPRD:03319|Vega:OTTHUMG00000150643 |
| Gene type | protein-coding |
| Map location | 5q35 |
| Pascal p-value | 0.086 |
| Sherlock p-value | 0.928 |
| Fetal beta | -1.559 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
| GSMA_IIA | Genome scan meta-analysis (All samples) | Psr: 0.0276 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0685 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg24421668 | 5 | 179244452 | SQSTM1 | 1.72E-8 | -0.023 | 6.24E-6 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs2450484 | chr5 | 179259841 | SQSTM1 | 8878 | 0.02 | cis | ||
| rs10277 | chr5 | 179264730 | SQSTM1 | 8878 | 0 | cis | ||
| rs1065154 | chr5 | 179264914 | SQSTM1 | 8878 | 0.04 | cis | ||
| rs748197 | chr5 | 179273819 | SQSTM1 | 8878 | 0.19 | cis | ||
| rs11249661 | chr5 | 179277873 | SQSTM1 | 8878 | 0.09 | cis | ||
| rs16827324 | chr1 | 40927958 | SQSTM1 | 8878 | 0 | trans | ||
| rs10277 | chr5 | 179264730 | SQSTM1 | 8878 | 0.2 | trans | ||
| rs1889151 | chr9 | 137160636 | SQSTM1 | 8878 | 0.12 | trans | ||
| rs7917302 | chr10 | 15474549 | SQSTM1 | 8878 | 0.03 | trans | ||
| rs10842557 | 0 | SQSTM1 | 8878 | 0.04 | trans | |||
| rs11161196 | chr15 | 25859201 | SQSTM1 | 8878 | 0.13 | trans |
Section II. Transcriptome annotation
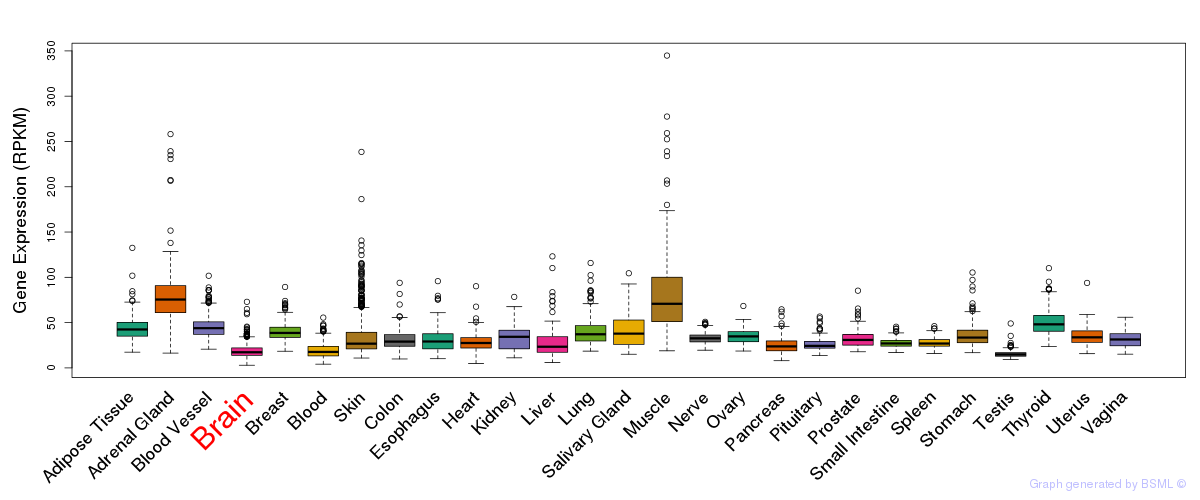
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| PINK1 | 0.89 | 0.89 |
| PRRT3 | 0.88 | 0.90 |
| CEND1 | 0.87 | 0.90 |
| TOM1 | 0.87 | 0.87 |
| DLGAP3 | 0.87 | 0.91 |
| TUBG2 | 0.87 | 0.91 |
| LENG3 | 0.86 | 0.86 |
| PPAPDC3 | 0.86 | 0.86 |
| ZFYVE28 | 0.85 | 0.89 |
| VGF | 0.85 | 0.91 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| TUBB2B | -0.57 | -0.56 |
| RBMX2 | -0.57 | -0.64 |
| CARHSP1 | -0.56 | -0.60 |
| TIGD1 | -0.56 | -0.47 |
| KIAA1949 | -0.56 | -0.45 |
| ZNF300 | -0.56 | -0.39 |
| C9orf46 | -0.56 | -0.62 |
| IGF2BP3 | -0.56 | -0.52 |
| TRAF4 | -0.56 | -0.60 |
| EXOSC8 | -0.56 | -0.57 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005515 | protein binding | IPI | 14676191 | |
| GO:0005515 | protein binding | ISS | - | |
| GO:0008270 | zinc ion binding | IEA | - | |
| GO:0042169 | SH2 domain binding | IDA | 8650207 | |
| GO:0042169 | SH2 domain binding | ISS | - | |
| GO:0030971 | receptor tyrosine kinase binding | TAS | 8650207 | |
| GO:0019901 | protein kinase binding | IDA | 8650207 | |
| GO:0019901 | protein kinase binding | ISS | - | |
| GO:0043130 | ubiquitin binding | IDA | 12857745 | |
| GO:0043130 | ubiquitin binding | ISS | - | |
| GO:0046872 | metal ion binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006511 | ubiquitin-dependent protein catabolic process | TAS | 8702753 | |
| GO:0007242 | intracellular signaling cascade | TAS | 8650207 | |
| GO:0006950 | response to stress | TAS | 12857745 | |
| GO:0008104 | protein localization | TAS | 8650207 | |
| GO:0006955 | immune response | IEA | - | |
| GO:0006915 | apoptosis | IEA | - | |
| GO:0016197 | endosome transport | TAS | 12857745 | |
| GO:0030154 | cell differentiation | IEA | - | |
| GO:0043122 | regulation of I-kappaB kinase/NF-kappaB cascade | IMP | 12857745 | |
| GO:0043122 | regulation of I-kappaB kinase/NF-kappaB cascade | ISS | - | |
| GO:0045944 | positive regulation of transcription from RNA polymerase II promoter | TAS | 12857745 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005829 | cytosol | NAS | - | |
| GO:0005829 | cytosol | TAS | 8650207 | |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0005770 | late endosome | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| C14orf139 | FLJ21276 | chromosome 14 open reading frame 139 | Two-hybrid | BioGRID | 16169070 |
| ERCC2 | COFS2 | EM9 | MGC102762 | MGC126218 | MGC126219 | TTD | XPD | excision repair cross-complementing rodent repair deficiency, complementation group 2 | - | HPRD | 8652557 |
| GABARAPL1 | APG8-LIKE | APG8L | ATG8 | ATG8L | GEC1 | GABA(A) receptor-associated protein like 1 | Two-hybrid | BioGRID | 16189514 |
| GABARAPL2 | ATG8 | GATE-16 | GATE16 | GEF-2 | GEF2 | GABA(A) receptor-associated protein-like 2 | Two-hybrid | BioGRID | 16189514 |
| IRAK1 | IRAK | pelle | interleukin-1 receptor-associated kinase 1 | - | HPRD | 12034707 |
| IRAK1 | IRAK | pelle | interleukin-1 receptor-associated kinase 1 | Affinity Capture-Western | BioGRID | 10747026 |
| LCK | YT16 | p56lck | pp58lck | lymphocyte-specific protein tyrosine kinase | - | HPRD,BioGRID | 8650207 |
| MAP1LC3B | LC3B | MAP1A/1BLC3 | microtubule-associated protein 1 light chain 3 beta | Two-hybrid | BioGRID | 16169070 |
| MAP2K5 | HsT17454 | MAPKK5 | MEK5 | PRKMK5 | mitogen-activated protein kinase kinase 5 | MEK5 interacts with p62. This interaction was modelled on a demonstrated interaction between mouse MEK5 and human p62. | BIND | 12813044 |
| NR2F2 | ARP1 | COUP-TFII | COUPTFB | MGC117452 | SVP40 | TFCOUP2 | nuclear receptor subfamily 2, group F, member 2 | - | HPRD | 8910285 |
| NTRK1 | DKFZp781I14186 | MTC | TRK | TRK1 | TRKA | p140-TrkA | neurotrophic tyrosine kinase, receptor, type 1 | Affinity Capture-Western | BioGRID | 11244088 |12471037 |
| NTRK2 | GP145-TrkB | TRKB | neurotrophic tyrosine kinase, receptor, type 2 | - | HPRD,BioGRID | 12471037 |
| NTRK3 | TRKC | gp145(trkC) | neurotrophic tyrosine kinase, receptor, type 3 | - | HPRD,BioGRID | 12471037 |
| PRKCI | DXS1179E | MGC26534 | PKCI | nPKC-iota | protein kinase C, iota | - | HPRD,BioGRID | 9566925 |
| PRKCI | DXS1179E | MGC26534 | PKCI | nPKC-iota | protein kinase C, iota | lamdaPKC interacts with p62. This interaction was modelled on a demonstrated interaction between mouse lamdaPKC and human p62. | BIND | 12813044 |
| PRKCZ | PKC-ZETA | PKC2 | protein kinase C, zeta | zetaPKC interacts with p62. This interaction was modelled on a demonstrated interaction between rat zetaPKC and human p62. | BIND | 12813044 |
| PRKCZ | PKC-ZETA | PKC2 | protein kinase C, zeta | - | HPRD | 9566925 |10356400 |
| PRKCZ | PKC-ZETA | PKC2 | protein kinase C, zeta | Affinity Capture-Western | BioGRID | 10747026 |
| PTPRJ | CD148 | DEP1 | HPTPeta | R-PTP-ETA | SCC1 | protein tyrosine phosphatase, receptor type, J | - | HPRD | 9115287 |
| RAD23A | HHR23A | MGC111083 | RAD23 homolog A (S. cerevisiae) | Affinity Capture-Western Two-hybrid | BioGRID | 16189514 |
| RIPK1 | FLJ39204 | RIP | RIP1 | receptor (TNFRSF)-interacting serine-threonine kinase 1 | - | HPRD,BioGRID | 10356400 |
| SNCA | MGC110988 | NACP | PARK1 | PARK4 | PD1 | synuclein, alpha (non A4 component of amyloid precursor) | - | HPRD | 11447312 |
| SQSTM1 | A170 | OSIL | PDB3 | ZIP3 | p60 | p62 | p62B | sequestosome 1 | p62 forms a dimer. | BIND | 12813044 |
| SQSTM1 | A170 | OSIL | PDB3 | ZIP3 | p60 | p62 | p62B | sequestosome 1 | Two-hybrid | BioGRID | 16169070 |
| TRAF6 | MGC:3310 | RNF85 | TNF receptor-associated factor 6 | - | HPRD,BioGRID | 10747026 |11244088 |
| TTN | CMD1G | CMH9 | CMPD4 | CONNECTIN | DKFZp451N061 | EOMFC | FLJ26020 | FLJ26409 | FLJ32040 | FLJ34413 | FLJ39564 | FLJ43066 | HMERF | LGMD2J | TMD | titin | Titin interacts with p62. This interaction was modelled on a demonstrated interaction between human Titin and rat p62. | BIND | 15802564 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PID IL1 PATHWAY | 34 | 28 | All SZGR 2.0 genes in this pathway |
| PID TNF PATHWAY | 46 | 33 | All SZGR 2.0 genes in this pathway |
| PID P75 NTR PATHWAY | 69 | 51 | All SZGR 2.0 genes in this pathway |
| PID TRKR PATHWAY | 62 | 48 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALLING BY NGF | 217 | 167 | All SZGR 2.0 genes in this pathway |
| REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | 12 | 8 | All SZGR 2.0 genes in this pathway |
| REACTOME P75NTR SIGNALS VIA NFKB | 14 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME NRIF SIGNALS CELL DEATH FROM THE NUCLEUS | 15 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | 11 | 8 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL DEATH SIGNALLING VIA NRAGE NRIF AND NADE | 60 | 43 | All SZGR 2.0 genes in this pathway |
| REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | 81 | 61 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY ILS | 107 | 86 | All SZGR 2.0 genes in this pathway |
| REACTOME IL1 SIGNALING | 39 | 30 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME CYTOKINE SIGNALING IN IMMUNE SYSTEM | 270 | 204 | All SZGR 2.0 genes in this pathway |
| GARY CD5 TARGETS UP | 473 | 314 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE LIVE UP | 485 | 293 | All SZGR 2.0 genes in this pathway |
| THUM SYSTOLIC HEART FAILURE DN | 244 | 147 | All SZGR 2.0 genes in this pathway |
| CASORELLI ACUTE PROMYELOCYTIC LEUKEMIA DN | 663 | 425 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| GARGALOVIC RESPONSE TO OXIDIZED PHOSPHOLIPIDS TURQUOISE UP | 79 | 50 | All SZGR 2.0 genes in this pathway |
| ODONNELL TFRC TARGETS UP | 456 | 228 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 TARGETS UP | 457 | 269 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC3 TARGETS UP | 501 | 327 | All SZGR 2.0 genes in this pathway |
| HUMMEL BURKITTS LYMPHOMA DN | 15 | 10 | All SZGR 2.0 genes in this pathway |
| TIEN INTESTINE PROBIOTICS 24HR DN | 214 | 133 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA REVERSIBLY UP | 8 | 6 | All SZGR 2.0 genes in this pathway |
| MARKEY RB1 CHRONIC LOF DN | 118 | 78 | All SZGR 2.0 genes in this pathway |
| CONCANNON APOPTOSIS BY EPOXOMICIN UP | 239 | 157 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA DN | 394 | 258 | All SZGR 2.0 genes in this pathway |
| MISSIAGLIA REGULATED BY METHYLATION UP | 126 | 78 | All SZGR 2.0 genes in this pathway |
| RASHI RESPONSE TO IONIZING RADIATION 2 | 127 | 92 | All SZGR 2.0 genes in this pathway |
| HERNANDEZ MITOTIC ARREST BY DOCETAXEL 2 UP | 64 | 45 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 UP | 309 | 199 | All SZGR 2.0 genes in this pathway |
| DIRMEIER LMP1 RESPONSE LATE UP | 57 | 41 | All SZGR 2.0 genes in this pathway |
| MOHANKUMAR TLX1 TARGETS UP | 414 | 287 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN DN | 584 | 395 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS UP | 1037 | 673 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS UP | 811 | 508 | All SZGR 2.0 genes in this pathway |
| FERREIRA EWINGS SARCOMA UNSTABLE VS STABLE DN | 98 | 59 | All SZGR 2.0 genes in this pathway |
| NAKAYAMA FRA2 TARGETS | 43 | 27 | All SZGR 2.0 genes in this pathway |
| SUH COEXPRESSED WITH ID1 AND ID2 UP | 19 | 14 | All SZGR 2.0 genes in this pathway |
| COLIN PILOCYTIC ASTROCYTOMA VS GLIOBLASTOMA DN | 28 | 21 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC TARGETS WITH EBOX | 230 | 156 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS | 775 | 494 | All SZGR 2.0 genes in this pathway |
| ZHANG RESPONSE TO IKK INHIBITOR AND TNF UP | 223 | 140 | All SZGR 2.0 genes in this pathway |
| UEDA PERIFERAL CLOCK | 169 | 111 | All SZGR 2.0 genes in this pathway |
| SHEPARD BMYB MORPHOLINO UP | 205 | 126 | All SZGR 2.0 genes in this pathway |
| GOLUB ALL VS AML DN | 24 | 14 | All SZGR 2.0 genes in this pathway |
| GALINDO IMMUNE RESPONSE TO ENTEROTOXIN | 85 | 67 | All SZGR 2.0 genes in this pathway |
| ASTIER INTEGRIN SIGNALING | 59 | 44 | All SZGR 2.0 genes in this pathway |
| NEMETH INFLAMMATORY RESPONSE LPS UP | 88 | 64 | All SZGR 2.0 genes in this pathway |
| BRUNO HEMATOPOIESIS | 66 | 48 | All SZGR 2.0 genes in this pathway |
| CHIBA RESPONSE TO TSA | 50 | 31 | All SZGR 2.0 genes in this pathway |
| SESTO RESPONSE TO UV C0 | 107 | 72 | All SZGR 2.0 genes in this pathway |
| GERHOLD ADIPOGENESIS UP | 49 | 40 | All SZGR 2.0 genes in this pathway |
| DOUGLAS BMI1 TARGETS DN | 314 | 188 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR DN | 911 | 527 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR UP | 783 | 442 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS SILENCED BY METHYLATION UP | 461 | 298 | All SZGR 2.0 genes in this pathway |
| WONG PROTEASOME GENE MODULE | 49 | 35 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER WITH H3K9ME3 DN | 120 | 71 | All SZGR 2.0 genes in this pathway |
| MITSIADES RESPONSE TO APLIDIN UP | 439 | 257 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER ERBB2 UP | 147 | 83 | All SZGR 2.0 genes in this pathway |
| PODAR RESPONSE TO ADAPHOSTIN UP | 147 | 98 | All SZGR 2.0 genes in this pathway |
| BLUM RESPONSE TO SALIRASIB UP | 245 | 159 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO CSF2RB AND IL4 UP | 338 | 225 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF UP | 418 | 282 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF VS CSF2RB AND IL4 UP | 408 | 276 | All SZGR 2.0 genes in this pathway |
| HAN SATB1 TARGETS UP | 395 | 249 | All SZGR 2.0 genes in this pathway |
| HAN SATB1 TARGETS DN | 442 | 275 | All SZGR 2.0 genes in this pathway |
| HSIAO HOUSEKEEPING GENES | 389 | 245 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH T 8 21 TRANSLOCATION | 368 | 247 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| MARTENS BOUND BY PML RARA FUSION | 456 | 287 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 24HR DN | 505 | 328 | All SZGR 2.0 genes in this pathway |
| WIERENGA STAT5A TARGETS UP | 217 | 131 | All SZGR 2.0 genes in this pathway |
| WIERENGA STAT5A TARGETS GROUP2 | 60 | 38 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 UP | 430 | 288 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP B | 549 | 316 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE LATE | 1137 | 655 | All SZGR 2.0 genes in this pathway |
| KOHOUTEK CCNT1 TARGETS | 50 | 26 | All SZGR 2.0 genes in this pathway |
| HUANG GATA2 TARGETS UP | 149 | 96 | All SZGR 2.0 genes in this pathway |
| DELACROIX RARG BOUND MEF | 367 | 231 | All SZGR 2.0 genes in this pathway |
| PHONG TNF RESPONSE VIA P38 PARTIAL | 160 | 106 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-17-5p/20/93.mr/106/519.d | 649 | 656 | 1A,m8 | hsa-miR-17-5p | CAAAGUGCUUACAGUGCAGGUAGU |
| hsa-miR-20abrain | UAAAGUGCUUAUAGUGCAGGUAG | ||||
| hsa-miR-106a | AAAAGUGCUUACAGUGCAGGUAGC | ||||
| hsa-miR-106bSZ | UAAAGUGCUGACAGUGCAGAU | ||||
| hsa-miR-20bSZ | CAAAGUGCUCAUAGUGCAGGUAG | ||||
| hsa-miR-519d | CAAAGUGCCUCCCUUUAGAGUGU | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.