Gene Page: SLC9A3R1
Summary ?
| GeneID | 9368 |
| Symbol | SLC9A3R1 |
| Synonyms | EBP50|NHERF|NHERF-1|NHERF1|NPHLOP2 |
| Description | SLC9A3 regulator 1 |
| Reference | MIM:604990|HGNC:HGNC:11075|Ensembl:ENSG00000109062|HPRD:05406|Vega:OTTHUMG00000178863 |
| Gene type | protein-coding |
| Map location | 17q25.1 |
| Pascal p-value | 0.991 |
| Sherlock p-value | 0.868 |
| Fetal beta | -1.397 |
| DMG | 1 (# studies) |
| eGene | Cerebellar Hemisphere Cerebellum Myers' cis & trans Meta |
| Support | ION BALANCE CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| DNM:Fromer_2014 | Whole Exome Sequencing analysis | This study reported a WES study of 623 schizophrenia trios, reporting DNMs using genomic DNA. | |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0231 |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| SLC9A3R1 | chr17 | 72758193 | A | G | NM_004252 | p.162S>G | missense | Schizophrenia | DNM:Fromer_2014 |
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg02976539 | 17 | 72757302 | SLC9A3R1 | 2.465E-4 | -0.41 | 0.037 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs2384957 | chr17 | 72784629 | SLC9A3R1 | 9368 | 0 | cis | ||
| rs13397687 | chr2 | 123655344 | SLC9A3R1 | 9368 | 0.07 | trans | ||
| rs13414989 | chr2 | 204991337 | SLC9A3R1 | 9368 | 0.14 | trans | ||
| rs9313483 | chr5 | 169448229 | SLC9A3R1 | 9368 | 0.07 | trans | ||
| rs9313484 | chr5 | 169448319 | SLC9A3R1 | 9368 | 0 | trans | ||
| rs794099 | chr6 | 164690612 | SLC9A3R1 | 9368 | 0.13 | trans | ||
| rs12545427 | chr8 | 74954879 | SLC9A3R1 | 9368 | 0.07 | trans | ||
| rs6472816 | chr8 | 74961543 | SLC9A3R1 | 9368 | 0.16 | trans | ||
| rs11216730 | chr11 | 117918701 | SLC9A3R1 | 9368 | 0.15 | trans | ||
| rs17504394 | chr13 | 30646519 | SLC9A3R1 | 9368 | 0.15 | trans | ||
| rs2384957 | chr17 | 72784629 | SLC9A3R1 | 9368 | 0.12 | trans |
Section II. Transcriptome annotation
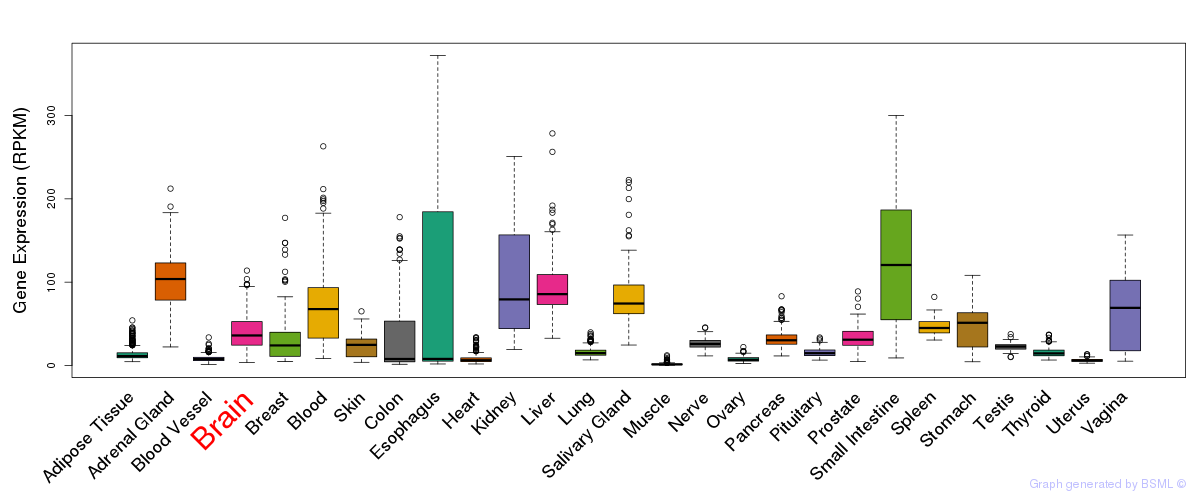
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| GYG2 | 0.69 | 0.65 |
| AC063977.1 | 0.66 | 0.72 |
| DNER | 0.63 | 0.71 |
| C15orf39 | 0.63 | 0.62 |
| ORAI2 | 0.63 | 0.76 |
| RADIL | 0.63 | 0.63 |
| MAP2K3 | 0.60 | 0.66 |
| LRRN2 | 0.60 | 0.72 |
| DCLK2 | 0.60 | 0.62 |
| CHST2 | 0.60 | 0.70 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.31 | -0.35 | -0.43 |
| AF347015.21 | -0.35 | -0.45 |
| MT-CO2 | -0.34 | -0.42 |
| AF347015.8 | -0.34 | -0.41 |
| AF347015.2 | -0.34 | -0.41 |
| AF347015.27 | -0.33 | -0.39 |
| AF347015.33 | -0.33 | -0.38 |
| MT-CYB | -0.32 | -0.39 |
| AF347015.15 | -0.31 | -0.37 |
| MT-ATP8 | -0.31 | -0.41 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005515 | protein binding | IPI | 11285285 |12471024 | |
| GO:0030165 | PDZ domain binding | IPI | 16236806 | |
| GO:0031698 | beta-2 adrenergic receptor binding | IPI | 9560162 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006461 | protein complex assembly | TAS | 9430655 | |
| GO:0016055 | Wnt receptor signaling pathway | IEA | - | |
| GO:0030643 | cellular phosphate ion homeostasis | IEA | - | |
| GO:0032415 | regulation of sodium:hydrogen antiporter activity | NAS | 9560162 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0042995 | cell projection | IEA | axon (GO term level: 4) | - |
| GO:0012505 | endomembrane system | IEA | - | |
| GO:0016020 | membrane | IEA | - | |
| GO:0015629 | actin cytoskeleton | TAS | 9430655 | |
| GO:0016324 | apical plasma membrane | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ABCC2 | ABC30 | CMOAT | DJS | KIAA1010 | MRP2 | cMRP | ATP-binding cassette, sub-family C (CFTR/MRP), member 2 | - | HPRD,BioGRID | 12615054 |
| ADRB2 | ADRB2R | ADRBR | B2AR | BAR | BETA2AR | adrenergic, beta-2-, receptor, surface | - | HPRD,BioGRID | 9560162 |9671706 |
| ADRB2 | ADRB2R | ADRBR | B2AR | BAR | BETA2AR | adrenergic, beta-2-, receptor, surface | PDZ domain of NHERF binds to the C-terminus of B2AR. | BIND | 9560162 |9671706 |12621035 |
| AKAP10 | D-AKAP2 | MGC9414 | PRKA10 | A kinase (PRKA) anchor protein 10 | - | HPRD | 14531806 |
| ATP6V1E1 | ATP6E | ATP6E2 | ATP6V1E | P31 | Vma4 | ATPase, H+ transporting, lysosomal 31kDa, V1 subunit E1 | - | HPRD | 10748165 |
| CFTR | ABC35 | ABCC7 | CF | CFTR/MRP | MRP7 | TNR-CFTR | dJ760C5.1 | cystic fibrosis transmembrane conductance regulator (ATP-binding cassette sub-family C, member 7) | First PDZ domain of EBP50 binds to the C-terminus of CFTR. | BIND | 9671706 |9677412 |10852925 |12621035 |
| CFTR | ABC35 | ABCC7 | CF | CFTR/MRP | MRP7 | TNR-CFTR | dJ760C5.1 | cystic fibrosis transmembrane conductance regulator (ATP-binding cassette sub-family C, member 7) | - | HPRD | 9613608 |9677412 |10852925 |
| CFTR | ABC35 | ABCC7 | CF | CFTR/MRP | MRP7 | TNR-CFTR | dJ760C5.1 | cystic fibrosis transmembrane conductance regulator (ATP-binding cassette sub-family C, member 7) | Affinity Capture-Western in vitro in vivo Reconstituted Complex | BioGRID | 9613608 |9671706 |9677412 |10852925 |12471024 |12615054 |
| CLCN3 | CLC3 | ClC-3 | chloride channel 3 | - | HPRD,BioGRID | 12471024 |
| CNGA2 | CNCA | CNCA1 | CNG2 | FLJ46312 | OCNC1 | OCNCALPHA | OCNCa | cyclic nucleotide gated channel alpha 2 | - | HPRD | 14604981 |
| CTNNB1 | CTNNB | DKFZp686D02253 | FLJ25606 | FLJ37923 | catenin (cadherin-associated protein), beta 1, 88kDa | - | HPRD,BioGRID | 12830000 |
| EZR | CVIL | CVL | DKFZp762H157 | FLJ26216 | MGC1584 | VIL2 | ezrin | - | HPRD,BioGRID | 9314537 |
| GNAQ | G-ALPHA-q | GAQ | guanine nucleotide binding protein (G protein), q polypeptide | - | HPRD,BioGRID | 12193606 |
| GNB2L1 | Gnb2-rs1 | H12.3 | HLC-7 | PIG21 | RACK1 | guanine nucleotide binding protein (G protein), beta polypeptide 2-like 1 | Affinity Capture-Western | BioGRID | 11956211 |
| KCNJ1 | KIR1.1 | ROMK | ROMK1 | potassium inwardly-rectifying channel, subfamily J, member 1 | - | HPRD,BioGRID | 14604981 |
| KCNJ1 | KIR1.1 | ROMK | ROMK1 | potassium inwardly-rectifying channel, subfamily J, member 1 | An unspecified isoform of ROMK interacts with NHERF-1. This interaction was modeled on a demonstrated interaction between Rat ROMK2 and Human NHERF-1. | BIND | 14604981 |
| LHCGR | FLJ41504 | LCGR | LGR2 | LH/CG-R | LH/CGR | LHR | LHRHR | LSH-R | luteinizing hormone/choriogonadotropin receptor | hLHR interacts with EBP50. | BIND | 14507927 |
| MSN | - | moesin | moesin interacts with hNHE-RF. | BIND | 9430655 |
| MSN | - | moesin | - | HPRD,BioGRID | 9430655 |
| NF2 | ACN | BANF | SCH | neurofibromin 2 (merlin) | - | HPRD,BioGRID | 9430655 |
| NF2 | ACN | BANF | SCH | neurofibromin 2 (merlin) | Merlin interacts with hNHE-RF | BIND | 9430655 |
| NOS2 | HEP-NOS | INOS | NOS | NOS2A | nitric oxide synthase 2, inducible | - | HPRD,BioGRID | 12080081 |
| OPRK1 | KOR | OPRK | opioid receptor, kappa 1 | - | HPRD,BioGRID | 12004055 |
| OPRK1 | KOR | OPRK | opioid receptor, kappa 1 | - | HPRD | 15070904 |
| P2RY1 | P2Y1 | purinergic receptor P2Y, G-protein coupled, 1 | NHERF interacts with P2Y1R. | BIND | 9671706 |
| P2RY1 | P2Y1 | purinergic receptor P2Y, G-protein coupled, 1 | - | HPRD,BioGRID | 9671706 |
| PAG1 | CBP | FLJ37858 | MGC138364 | PAG | phosphoprotein associated with glycosphingolipid microdomains 1 | - | HPRD,BioGRID | 11684085 |
| PDGFRA | CD140A | MGC74795 | PDGFR2 | Rhe-PDGFRA | platelet-derived growth factor receptor, alpha polypeptide | - | HPRD,BioGRID | 11046132 |11882663 |
| PDGFRB | CD140B | JTK12 | PDGF-R-beta | PDGFR | PDGFR1 | platelet-derived growth factor receptor, beta polypeptide | - | HPRD,BioGRID | 11046132 |
| PDZK1 | CAP70 | CLAMP | NHERF3 | PDZD1 | PDZ domain containing 1 | Reconstituted Complex Two-hybrid | BioGRID | 14531806 |
| PIK3CA | MGC142161 | MGC142163 | PI3K | p110-alpha | phosphoinositide-3-kinase, catalytic, alpha polypeptide | Reconstituted Complex | BioGRID | 12954600 |
| PLCB1 | FLJ45792 | PI-PLC | PLC-154 | PLC-I | PLC154 | phospholipase C, beta 1 (phosphoinositide-specific) | - | HPRD,BioGRID | 10980202 |
| PLCB2 | FLJ38135 | phospholipase C, beta 2 | - | HPRD | 10980202 |
| PTH1R | MGC138426 | MGC138452 | PTHR | PTHR1 | parathyroid hormone 1 receptor | - | HPRD,BioGRID | 12075354 |
| RDX | DFNB24 | radixin | Radixin interacts with hNHE-RF. This interaction was modeled on a demonstrated interaction between chicken radixin and human hNHE-RF. | BIND | 9430655 |
| RDX | DFNB24 | radixin | - | HPRD | 9430655|12499563 |
| SLC4A8 | DKFZp761B2318 | FLJ46462 | NBC3 | solute carrier family 4, sodium bicarbonate cotransporter, member 8 | - | HPRD,BioGRID | 12444018 |
| SLC9A3 | MGC126718 | MGC126720 | NHE3 | solute carrier family 9 (sodium/hydrogen exchanger), member 3 | - | HPRD,BioGRID | 14580213 |
| SLC9A3R1 | EBP50 | NHERF | NHERF1 | NPHLOP2 | solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 1 | Far Western Reconstituted Complex | BioGRID | 11046132 |15070904 |
| TBC1D10A | EPI64 | TBC1D10 | dJ130H16.1 | dJ130H16.2 | TBC1 domain family, member 10A | - | HPRD,BioGRID | 11285285 |
| TRPC5 | TRP5 | transient receptor potential cation channel, subfamily C, member 5 | - | HPRD | 10980202 |
| YAP1 | YAP | YAP2 | YAP65 | YKI | Yes-associated protein 1, 65kDa | - | HPRD,BioGRID | 10562288 |
| YES1 | HsT441 | P61-YES | Yes | c-yes | v-yes-1 Yamaguchi sarcoma viral oncogene homolog 1 | Reconstituted Complex | BioGRID | 10562288 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| BIOCARTA CFTR PATHWAY | 12 | 11 | All SZGR 2.0 genes in this pathway |
| WNT SIGNALING | 89 | 71 | All SZGR 2.0 genes in this pathway |
| PID TXA2PATHWAY | 57 | 43 | All SZGR 2.0 genes in this pathway |
| PID PDGFRB PATHWAY | 129 | 103 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA DN | 526 | 357 | All SZGR 2.0 genes in this pathway |
| ZHONG RESPONSE TO AZACITIDINE AND TSA UP | 183 | 119 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS BASAL UP | 380 | 215 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS MESENCHYMAL UP | 450 | 256 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN NPM1 MUTATED SIGNATURE 1 DN | 126 | 86 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN NPM1 MUTATED SIGNATURE 2 DN | 77 | 46 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN NPM1 SIGNATURE 3 DN | 162 | 116 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 1 DN | 378 | 231 | All SZGR 2.0 genes in this pathway |
| WANG ESOPHAGUS CANCER VS NORMAL DN | 101 | 66 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL TGFB1 TARGETS DN | 62 | 44 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER APOCRINE VS BASAL | 330 | 217 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER BASAL VS LULMINAL | 330 | 215 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE UP | 134 | 93 | All SZGR 2.0 genes in this pathway |
| PATIL LIVER CANCER | 747 | 453 | All SZGR 2.0 genes in this pathway |
| LIEN BREAST CARCINOMA METAPLASTIC VS DUCTAL DN | 114 | 58 | All SZGR 2.0 genes in this pathway |
| GROSS HYPOXIA VIA ELK3 UP | 209 | 139 | All SZGR 2.0 genes in this pathway |
| GROSS HYPOXIA VIA ELK3 AND HIF1A DN | 103 | 71 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS DN | 848 | 527 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| GOTZMANN EPITHELIAL TO MESENCHYMAL TRANSITION DN | 206 | 136 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 17Q21 Q25 AMPLICON | 335 | 181 | All SZGR 2.0 genes in this pathway |
| UEDA PERIFERAL CLOCK | 169 | 111 | All SZGR 2.0 genes in this pathway |
| STOSSI RESPONSE TO ESTRADIOL | 50 | 35 | All SZGR 2.0 genes in this pathway |
| LEI MYB TARGETS | 318 | 215 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 16HR UP | 225 | 139 | All SZGR 2.0 genes in this pathway |
| DEBIASI APOPTOSIS BY REOVIRUS INFECTION DN | 287 | 208 | All SZGR 2.0 genes in this pathway |
| TSENG IRS1 TARGETS UP | 113 | 71 | All SZGR 2.0 genes in this pathway |
| BILD E2F3 ONCOGENIC SIGNATURE | 246 | 153 | All SZGR 2.0 genes in this pathway |
| RIGGINS TAMOXIFEN RESISTANCE UP | 66 | 44 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER LUMINAL B UP | 172 | 109 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL DN | 701 | 446 | All SZGR 2.0 genes in this pathway |
| WEST ADRENOCORTICAL TUMOR DN | 546 | 362 | All SZGR 2.0 genes in this pathway |
| BLUM RESPONSE TO SALIRASIB DN | 342 | 220 | All SZGR 2.0 genes in this pathway |
| WANG TUMOR INVASIVENESS DN | 210 | 128 | All SZGR 2.0 genes in this pathway |
| VANTVEER BREAST CANCER ESR1 UP | 167 | 99 | All SZGR 2.0 genes in this pathway |
| LEE DIFFERENTIATING T LYMPHOCYTE | 200 | 115 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3 | 1069 | 729 | All SZGR 2.0 genes in this pathway |
| COULOUARN TEMPORAL TGFB1 SIGNATURE DN | 138 | 99 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH T 8 21 TRANSLOCATION | 368 | 247 | All SZGR 2.0 genes in this pathway |
| MARSON FOXP3 TARGETS UP | 66 | 43 | All SZGR 2.0 genes in this pathway |
| KARLSSON TGFB1 TARGETS DN | 207 | 139 | All SZGR 2.0 genes in this pathway |
| WANG RESPONSE TO GSK3 INHIBITOR SB216763 DN | 374 | 217 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 6HR UP | 229 | 149 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 24HR UP | 324 | 193 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| BHAT ESR1 TARGETS NOT VIA AKT1 UP | 211 | 131 | All SZGR 2.0 genes in this pathway |
| BHAT ESR1 TARGETS VIA AKT1 UP | 281 | 183 | All SZGR 2.0 genes in this pathway |
| VANDESLUIS COMMD1 TARGETS GROUP 3 UP | 89 | 50 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS UP | 682 | 433 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY STEM CELL DN | 428 | 246 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-339 | 404 | 410 | m8 | hsa-miR-339 | UCCCUGUCCUCCAGGAGCUCA |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.