Gene Page: MAPK8IP1
Summary ?
| GeneID | 9479 |
| Symbol | MAPK8IP1 |
| Synonyms | IB1|JIP-1|JIP1|PRKM8IP |
| Description | mitogen-activated protein kinase 8 interacting protein 1 |
| Reference | MIM:604641|HGNC:HGNC:6882|Ensembl:ENSG00000121653|HPRD:05223|Vega:OTTHUMG00000134324 |
| Gene type | protein-coding |
| Map location | 11p11.2 |
| Pascal p-value | 0.005 |
| Sherlock p-value | 0.999 |
| Fetal beta | -0.926 |
| DMG | 1 (# studies) |
| eGene | Cerebellar Hemisphere Putamen basal ganglia |
| Support | INTRACELLULAR SIGNAL TRANSDUCTION G2Cdb.humanNRC G2Cdb.humanPSP CompositeSet Darnell FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Nishioka_2013 | Genome-wide DNA methylation analysis | The authors investigated the methylation profiles of DNA in peripheral blood cells from 18 patients with first-episode schizophrenia (FESZ) and from 15 normal controls. | 1 |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.1391 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg03568064 | 11 | 45907500 | MAPK8IP1 | -0.023 | 0.39 | DMG:Nishioka_2013 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs7483388 | 11 | 45168327 | MAPK8IP1 | ENSG00000121653.7 | 2.62712E-6 | 0.04 | -738875 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
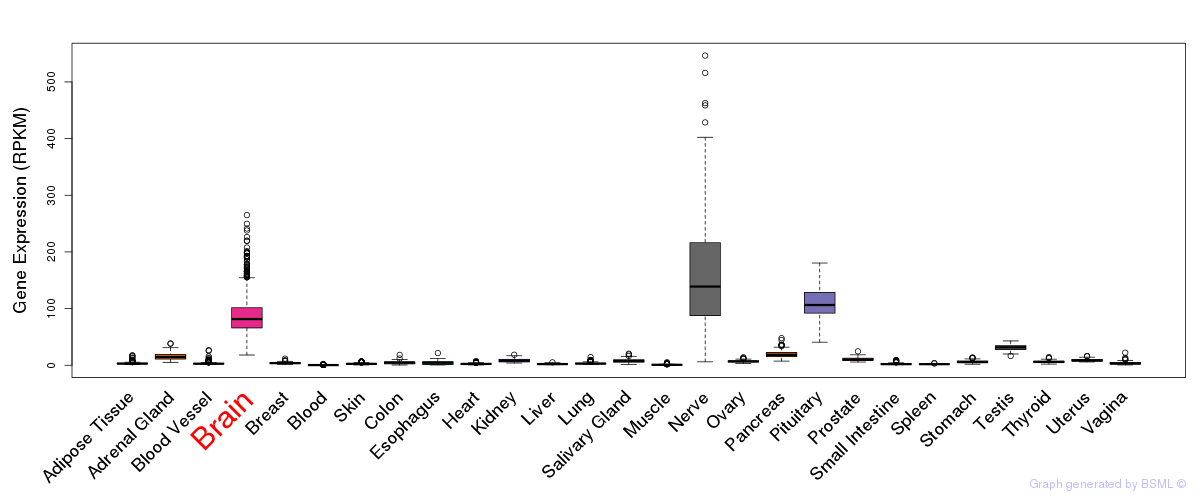
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005078 | MAP-kinase scaffold activity | IPI | 11238452 | |
| GO:0005515 | protein binding | IPI | 11726277 | |
| GO:0004860 | protein kinase inhibitor activity | TAS | 9933567 | |
| GO:0019894 | kinesin binding | IPI | 11238452 | |
| GO:0016301 | kinase activity | IEA | - | |
| GO:0019901 | protein kinase binding | IPI | 11238452 |11726277 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0016192 | vesicle-mediated transport | ISS | - | |
| GO:0046328 | regulation of JNK cascade | ISS | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005737 | cytoplasm | IDA | 10574993 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| AKT1 | AKT | MGC99656 | PKB | PKB-ALPHA | PRKBA | RAC | RAC-ALPHA | v-akt murine thymoma viral oncogene homolog 1 | - | HPRD,BioGRID | 12194869 |
| APP | AAA | ABETA | ABPP | AD1 | APPI | CTFgamma | CVAP | PN2 | amyloid beta (A4) precursor protein | JIP1 interacts with APP. | BIND | 11724784 |
| APP | AAA | ABETA | ABPP | AD1 | APPI | CTFgamma | CVAP | PN2 | amyloid beta (A4) precursor protein | - | HPRD | 11517249 |11724784 |
| DUSP16 | KIAA1700 | MGC129701 | MGC129702 | MKP-7 | MKP7 | dual specificity phosphatase 16 | - | HPRD,BioGRID | 12524447 |
| LRP1 | A2MR | APOER | APR | CD91 | FLJ16451 | IGFBP3R | LRP | MGC88725 | TGFBR5 | low density lipoprotein-related protein 1 (alpha-2-macroglobulin receptor) | - | HPRD,BioGRID | 10827173 |
| LRP2 | DBS | gp330 | low density lipoprotein-related protein 2 | - | HPRD,BioGRID | 10827173 |
| LRP8 | APOER2 | HSZ75190 | MCI1 | low density lipoprotein receptor-related protein 8, apolipoprotein e receptor | Two-hybrid | BioGRID | 10827173 |
| MAP2K7 | Jnkk2 | MAPKK7 | MKK7 | PRKMK7 | mitogen-activated protein kinase kinase 7 | - | HPRD,BioGRID | 10490659 |
| MAP3K10 | MLK2 | MST | mitogen-activated protein kinase kinase kinase 10 | Affinity Capture-Western Reconstituted Complex | BioGRID | 10490659 |
| MAP3K11 | MGC17114 | MLK-3 | MLK3 | PTK1 | SPRK | mitogen-activated protein kinase kinase kinase 11 | Affinity Capture-Western Reconstituted Complex | BioGRID | 10490659 |
| MAP3K12 | DLK | MUK | ZPK | ZPKP1 | mitogen-activated protein kinase kinase kinase 12 | Affinity Capture-Western Reconstituted Complex | BioGRID | 10490659 |
| MAP3K13 | LZK | MGC133196 | mitogen-activated protein kinase kinase kinase 13 | - | HPRD,BioGRID | 11726277 |
| MAPK8 | JNK | JNK1 | JNK1A2 | JNK21B1/2 | PRKM8 | SAPK1 | mitogen-activated protein kinase 8 | - | HPRD,BioGRID | 9733513 |
| MAPK8IP1 | IB1 | JIP-1 | JIP1 | PRKM8IP | mitogen-activated protein kinase 8 interacting protein 1 | Affinity Capture-Western | BioGRID | 10490659 |
| MAPK8IP2 | IB2 | JIP2 | PRKM8IPL | mitogen-activated protein kinase 8 interacting protein 2 | Affinity Capture-Western Reconstituted Complex | BioGRID | 10490659 |
| MAPK8IP3 | DKFZp762N1113 | FLJ00027 | JIP3 | JSAP1 | KIAA1066 | SYD2 | mitogen-activated protein kinase 8 interacting protein 3 | - | HPRD,BioGRID | 10629060 |
| MAPK9 | JNK-55 | JNK2 | JNK2A | JNK2ALPHA | JNK2B | JNK2BETA | PRKM9 | SAPK | p54a | p54aSAPK | mitogen-activated protein kinase 9 | Affinity Capture-Western Reconstituted Complex | BioGRID | 9733513 |10490659 |
| PAX2 | - | paired box 2 | Affinity Capture-Western | BioGRID | 11700324 |
| RECQL5 | FLJ90603 | RECQ5 | RecQ protein-like 5 | Two-hybrid | BioGRID | 16169070 |
| RGNEF | DKFZp686P12164 | FLJ21817 | KIAA1998 | Rho-guanine nucleotide exchange factor | - | HPRD | 10574993 |
| SNCA | MGC110988 | NACP | PARK1 | PARK4 | PD1 | synuclein, alpha (non A4 component of amyloid precursor) | - | HPRD | 11790792 |
| TIAM1 | FLJ36302 | T-cell lymphoma invasion and metastasis 1 | - | HPRD,BioGRID | 12024021 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG MAPK SIGNALING PATHWAY | 267 | 205 | All SZGR 2.0 genes in this pathway |
| ST DIFFERENTIATION PATHWAY IN PC12 CELLS | 45 | 35 | All SZGR 2.0 genes in this pathway |
| SIG CD40PATHWAYMAP | 34 | 28 | All SZGR 2.0 genes in this pathway |
| ST GRANULE CELL SURVIVAL PATHWAY | 27 | 23 | All SZGR 2.0 genes in this pathway |
| ST INTEGRIN SIGNALING PATHWAY | 82 | 62 | All SZGR 2.0 genes in this pathway |
| ST FAS SIGNALING PATHWAY | 65 | 54 | All SZGR 2.0 genes in this pathway |
| PID REELIN PATHWAY | 29 | 29 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC3 TARGETS UP | 501 | 327 | All SZGR 2.0 genes in this pathway |
| LANDIS ERBB2 BREAST TUMORS 324 UP | 150 | 93 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 3 4WK UP | 214 | 144 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 4 5WK DN | 196 | 131 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 6 7WK UP | 197 | 135 | All SZGR 2.0 genes in this pathway |
| RAMJAUN APOPTOSIS BY TGFB1 VIA SMAD4 DN | 8 | 6 | All SZGR 2.0 genes in this pathway |
| ROVERSI GLIOMA COPY NUMBER UP | 100 | 75 | All SZGR 2.0 genes in this pathway |
| SNIJDERS AMPLIFIED IN HEAD AND NECK TUMORS | 37 | 27 | All SZGR 2.0 genes in this pathway |
| KANG GIST WITH PDGFRA UP | 50 | 27 | All SZGR 2.0 genes in this pathway |
| SHETH LIVER CANCER VS TXNIP LOSS PAM1 | 229 | 137 | All SZGR 2.0 genes in this pathway |
| GOERING BLOOD HDL CHOLESTEROL QTL TRANS | 14 | 7 | All SZGR 2.0 genes in this pathway |
| LE EGR2 TARGETS DN | 108 | 84 | All SZGR 2.0 genes in this pathway |
| NING CHRONIC OBSTRUCTIVE PULMONARY DISEASE UP | 157 | 105 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| ULE SPLICING VIA NOVA2 | 43 | 38 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS UP | 317 | 208 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS SILENCED BY METHYLATION UP | 461 | 298 | All SZGR 2.0 genes in this pathway |
| MCCABE BOUND BY HOXC6 | 469 | 239 | All SZGR 2.0 genes in this pathway |
| RAY TUMORIGENESIS BY ERBB2 CDC25A DN | 159 | 105 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS 36HR UP | 221 | 150 | All SZGR 2.0 genes in this pathway |
| JISON SICKLE CELL DISEASE DN | 181 | 97 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3K4ME2 | 491 | 319 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP B | 549 | 316 | All SZGR 2.0 genes in this pathway |
| LIU IL13 MEMORY MODEL UP | 17 | 12 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-183 | 726 | 732 | m8 | hsa-miR-183 | UAUGGCACUGGUAGAAUUCACUG |
| miR-214 | 58 | 64 | 1A | hsa-miR-214brain | ACAGCAGGCACAGACAGGCAG |
| miR-217 | 171 | 177 | m8 | hsa-miR-217 | UACUGCAUCAGGAACUGAUUGGAU |
| miR-24 | 460 | 466 | m8 | hsa-miR-24SZ | UGGCUCAGUUCAGCAGGAACAG |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.