Gene Page: POLR1C
Summary ?
| GeneID | 9533 |
| Symbol | POLR1C |
| Synonyms | AC40|HLD11|RPA39|RPA40|RPA5|RPAC1|RPC40|TCS3 |
| Description | polymerase (RNA) I subunit C |
| Reference | MIM:610060|HGNC:HGNC:20194|Ensembl:ENSG00000171453|HPRD:09661|Vega:OTTHUMG00000014739 |
| Gene type | protein-coding |
| Map location | 6p21.1 |
| Pascal p-value | 3.739E-5 |
| Sherlock p-value | 0.454 |
| Fetal beta | 0.225 |
| DMG | 1 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
| GSMA_IIE | Genome scan meta-analysis (European-ancestry samples) | Psr: 0.04433 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg14490972 | 6 | 43484812 | POLR1C | 8.75E-10 | -0.019 | 1.09E-6 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
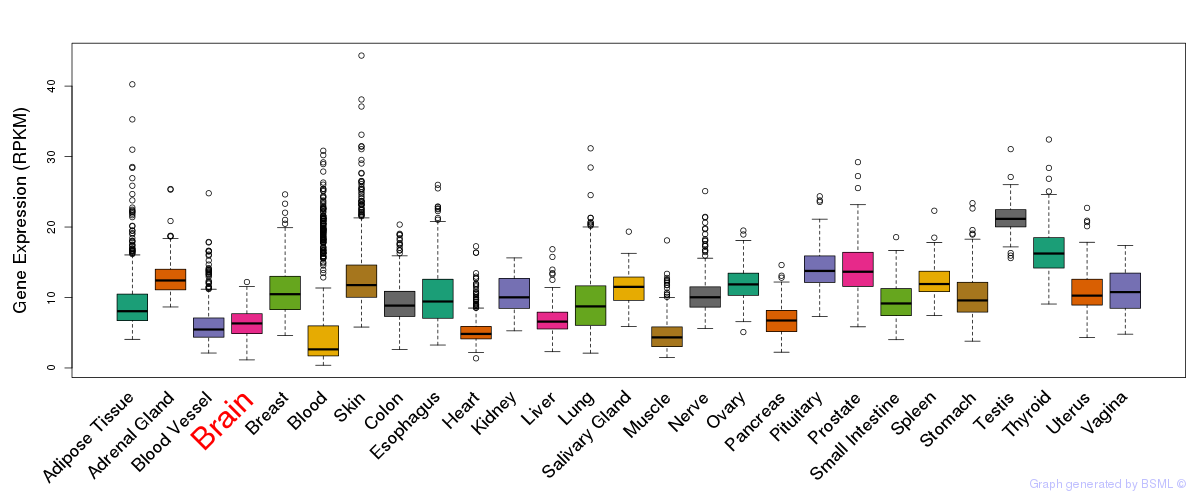
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| CNTNAP1 | 0.93 | 0.88 |
| KCNJ9 | 0.91 | 0.88 |
| GABARAPL3 | 0.90 | 0.89 |
| NPTN | 0.90 | 0.87 |
| CAMTA2 | 0.90 | 0.92 |
| EHD3 | 0.90 | 0.87 |
| SYNGR1 | 0.90 | 0.85 |
| SLC12A5 | 0.90 | 0.90 |
| RPH3A | 0.89 | 0.82 |
| PTPRN | 0.89 | 0.89 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| BCL7C | -0.54 | -0.62 |
| C9orf46 | -0.49 | -0.50 |
| RPL23A | -0.47 | -0.53 |
| RPL35 | -0.47 | -0.54 |
| RPL18 | -0.47 | -0.55 |
| RPS19P3 | -0.47 | -0.57 |
| RPS18 | -0.46 | -0.48 |
| RPS8 | -0.46 | -0.48 |
| RPLP1 | -0.46 | -0.50 |
| RPS6 | -0.46 | -0.49 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003677 | DNA binding | IEA | - | |
| GO:0003899 | DNA-directed RNA polymerase activity | IEA | - | |
| GO:0003899 | DNA-directed RNA polymerase activity | TAS | 9540830 | |
| GO:0046983 | protein dimerization activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006350 | transcription | IEA | - | |
| GO:0006360 | transcription from RNA polymerase I promoter | TAS | 9540830 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005654 | nucleoplasm | EXP | 9582279 |12393749 |12646563 | |
| GO:0005736 | DNA-directed RNA polymerase I complex | TAS | 9540830 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG PURINE METABOLISM | 159 | 96 | All SZGR 2.0 genes in this pathway |
| KEGG PYRIMIDINE METABOLISM | 98 | 53 | All SZGR 2.0 genes in this pathway |
| KEGG RNA POLYMERASE | 29 | 12 | All SZGR 2.0 genes in this pathway |
| KEGG CYTOSOLIC DNA SENSING PATHWAY | 56 | 44 | All SZGR 2.0 genes in this pathway |
| BIOCARTA ARF PATHWAY | 17 | 13 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | 23 | 12 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL I TRANSCRIPTION TERMINATION | 22 | 12 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL I TRANSCRIPTION | 89 | 64 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL III TRANSCRIPTION | 33 | 18 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSCRIPTION | 210 | 127 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL I RNA POL III AND MITOCHONDRIAL TRANSCRIPTION | 122 | 80 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | 26 | 13 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL III TRANSCRIPTION TERMINATION | 19 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL III CHAIN ELONGATION | 17 | 8 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL I TRANSCRIPTION INITIATION | 25 | 14 | All SZGR 2.0 genes in this pathway |
| GARY CD5 TARGETS DN | 431 | 263 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| VANHARANTA UTERINE FIBROID WITH 7Q DELETION DN | 36 | 23 | All SZGR 2.0 genes in this pathway |
| GRASEMANN RETINOBLASTOMA WITH 6P AMPLIFICATION | 14 | 14 | All SZGR 2.0 genes in this pathway |
| AMUNDSON RESPONSE TO ARSENITE | 217 | 143 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| KOYAMA SEMA3B TARGETS DN | 411 | 249 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS | 775 | 494 | All SZGR 2.0 genes in this pathway |
| STARK PREFRONTAL CORTEX 22Q11 DELETION DN | 517 | 309 | All SZGR 2.0 genes in this pathway |
| SHEPARD BMYB MORPHOLINO UP | 205 | 126 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| FLECHNER PBL KIDNEY TRANSPLANT REJECTED VS OK UP | 63 | 48 | All SZGR 2.0 genes in this pathway |
| MAHAJAN RESPONSE TO IL1A DN | 76 | 57 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| MODY HIPPOCAMPUS PRENATAL | 42 | 33 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR DN | 911 | 527 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR DN | 1011 | 592 | All SZGR 2.0 genes in this pathway |
| MARZEC IL2 SIGNALING UP | 115 | 80 | All SZGR 2.0 genes in this pathway |
| ZHANG BREAST CANCER PROGENITORS UP | 425 | 253 | All SZGR 2.0 genes in this pathway |
| MUELLER PLURINET | 299 | 189 | All SZGR 2.0 genes in this pathway |
| WANG TUMOR INVASIVENESS UP | 374 | 247 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS UP | 491 | 316 | All SZGR 2.0 genes in this pathway |
| HAN SATB1 TARGETS UP | 395 | 249 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA CLASSES UP | 605 | 377 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE DN | 841 | 431 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 2 DN | 336 | 211 | All SZGR 2.0 genes in this pathway |
| BILANGES RAPAMYCIN SENSITIVE VIA TSC1 AND TSC2 | 73 | 37 | All SZGR 2.0 genes in this pathway |