|
|
||
|
|
||
Data collection of TSGene and how to use TSGene: |
|---|
|
1. Data collection of TSGene database Four data sources for tumor suppressor genes Curation tumor suppressor genes from literature 2. Information for tumor suppressor genes General information and literature evidence Text search of tumor suppressor gene Quick access information in database Blast all protein and nucleotide sequences By data sources, chromosome, and gene type |
Data collection of TSGene database |
|
The primary aim of the database is to support cancer research by maintaining a high quality tumor suppressor gene database that serves as a comprehensive, fully classified, richly and accurately annotated tumor suppressor gene knowledgebase, with extensive cross-references and querying interfaces freely accessible to the scientific community. |
Four data sources for tumor suppressor genes |
|
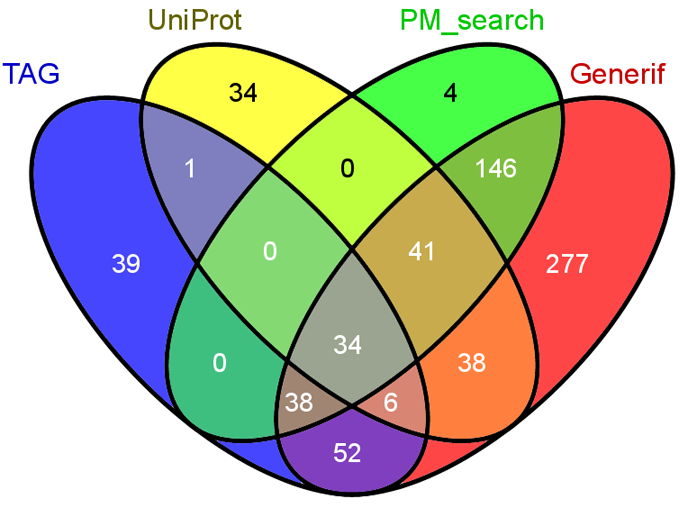
Construction of this tumor suppressor gene database for Human, Mouse, and Rat genes involved four main steps: integration of tumor suppressor genes from UniProt keyword "Tumor suppressor" and Tumor Associated Gene (TAG) database; curation of tumor suppressor genes from published literatures; mapping the description of tumor suppressor genes from literatures to Entrez gene database IDs; manual curation of each literature for its organism information; extensive annotations of cellular function, gene expression, mutation, methylation, transcription factor, post-translational modification, and protein-protein interaction. The gene contents from the two databases and two literature sources were summarized in the figure below. The detail for the two literature sources is in the following section. |
| Curation of tumor suppressor genes from literature [ top ] |
Curation about tumor suppressor genes from the literature includes five steps before finally being included in the TSGene database: exhaustive searching for relevant abstracts from the PubMed and Generif databases using the key words "tumor suppressor gene;" extracting the description for the tumor suppressor gene from text; grouping the descriptions extracted from PubMed abstracts and Generfi records by their topics using Entrez related topic function; extraction of gene name from the grouped descriptions of the tumor suppressor genes; lastly, mapping the gene name to Entrez geneID. Exhaustive search: Extracting description: Group abstracts: Curation: Mappping the gene symbols: |
Information for tumor suppressor genes [ top ] |
|
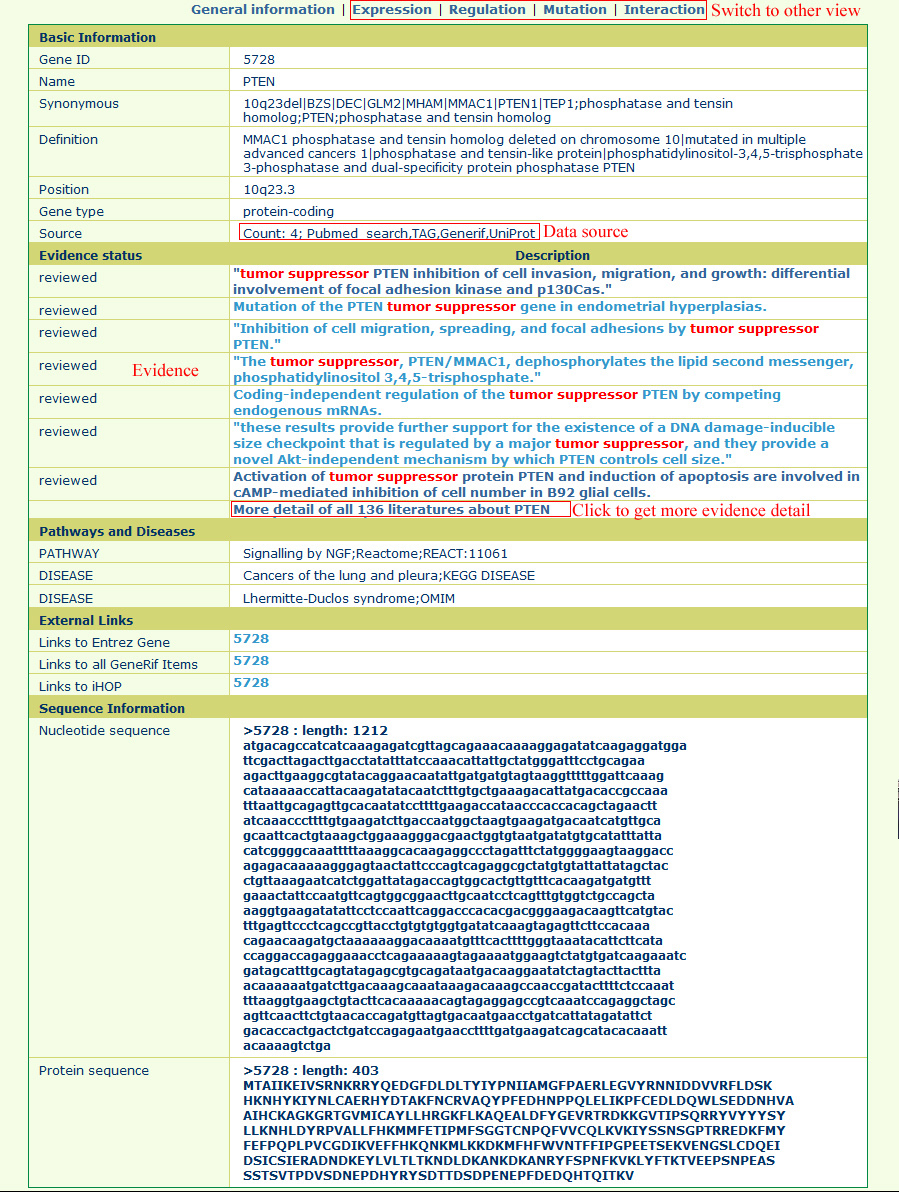
Information is represented on six different types of pages, including general information view, literature highlight view, gene expression view, gene regulation view, gene mutation view, and gene interaction view. The general information page is like the following:In this page, users can find the data source and our curated descriptions for tumor suppressor genes from literature. It is easy to switch to other annotations by clicking the hyperlink at the top of the page.
User can find the details of the literatures with keywords highlighted in the literature highlight page as below. The keyword "tumor suppressor" is marked in red; keywords such as "cancer" and "pathway" are marked in brown; and the keywords in the category of "microRNA" are highlighted in green; the keywords such as "mutation" and "expression" are marked in black as shown in below.
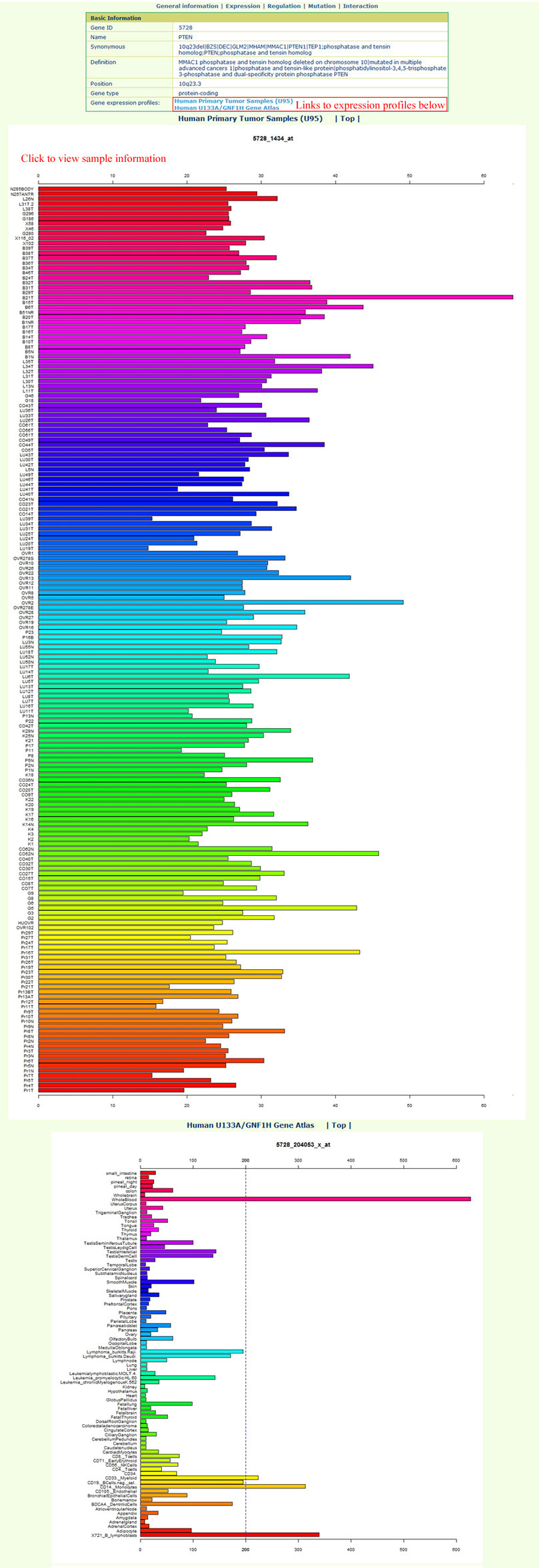
The gene expression page is as below:In this page, users can find gene expression profiles from 184 human tumor samples and 84 normal tissue samples from BioGPS. It is easy to view the sample information of the 184 tumor samples by clicking the hyperlink in the profile images. Some genes have multiple probes; to provide an unbiased view for users, we presented all the gene expressions from all probes without any modification.
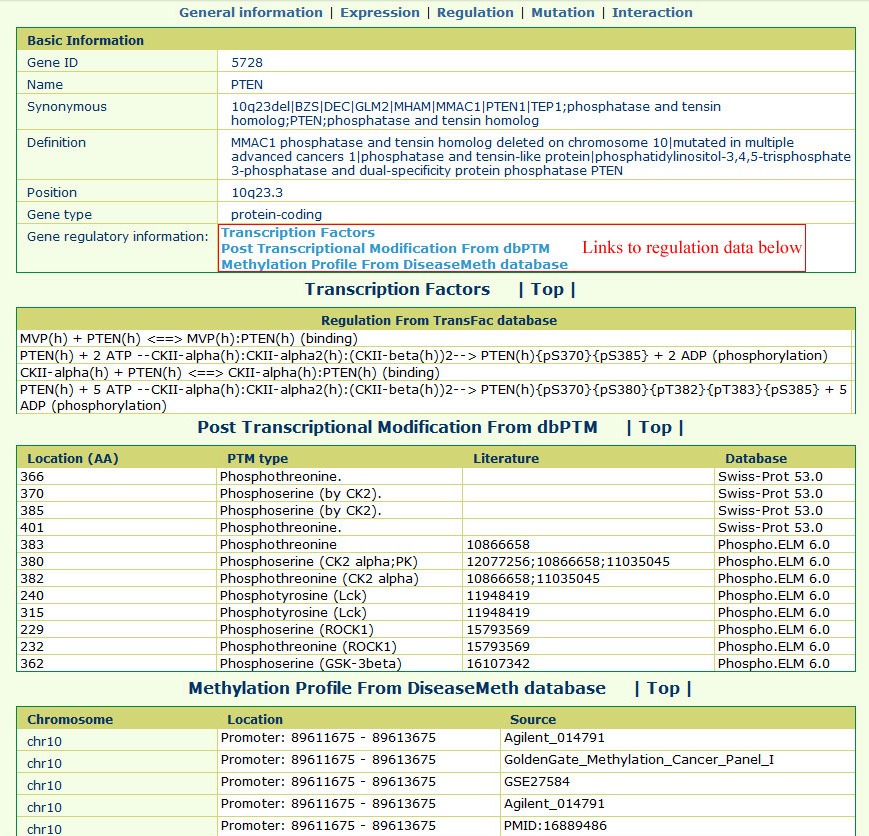
The gene regulation page appears as follows:The transcription factor regulation and post-transcriptional modification information were integrated from the TRANSFAC and dbPTM databases. In addition, the methylation in promoter regions was annotated based on data from the DiseaseMeth database.
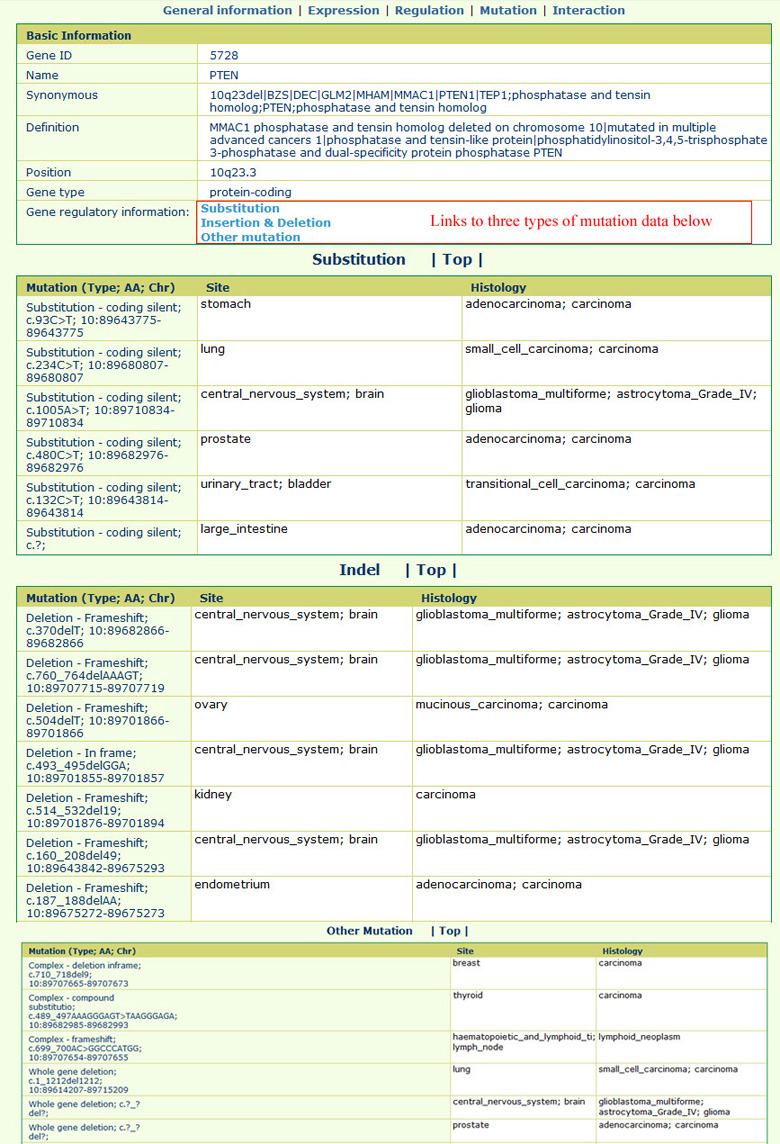
The gene mutation page appears as follows:All the related mutations were collected from the COSMIC database; we further divided the mutations into three main types, including "Substitution," "Insertion & Deletion," and "Other mutation."
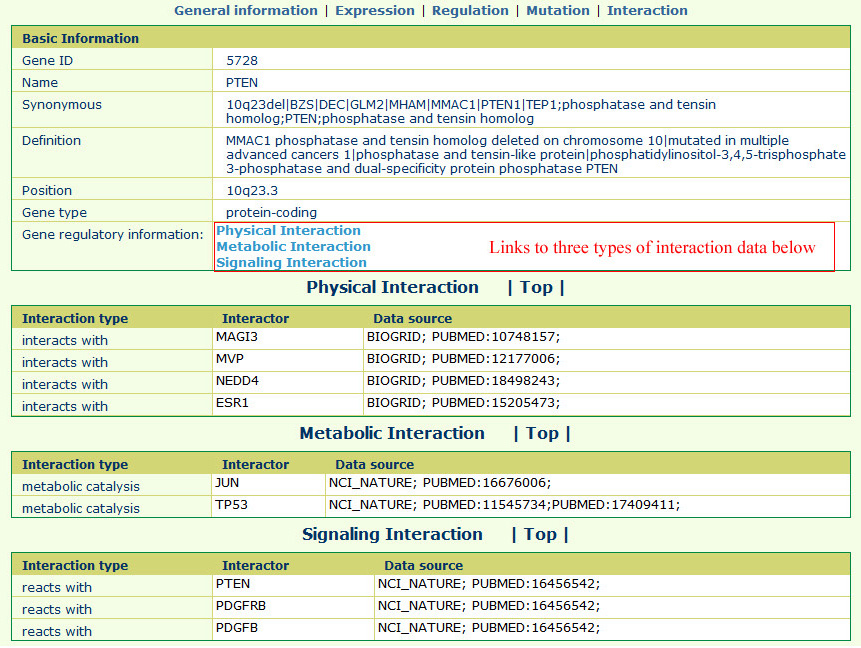
The gene interaction page appears as follows:All the related protein-protein interactions were collected from the PathwayCommon database; we further divided the interactions into three main types, including "Physical Interaction," "Metabolic Interaction," and "Signaling Interaction."
|
|
|
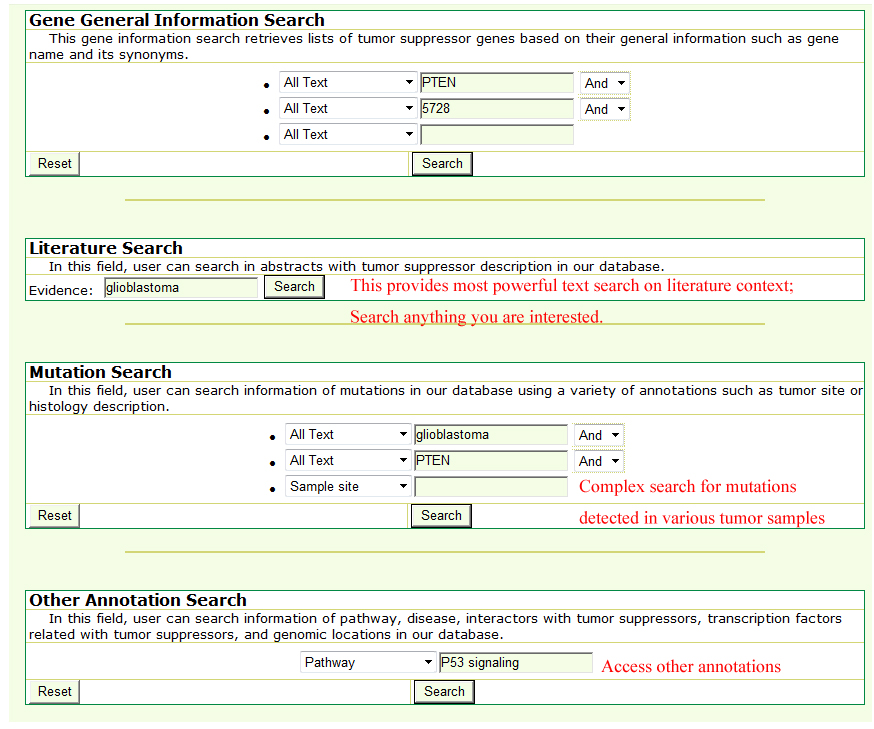
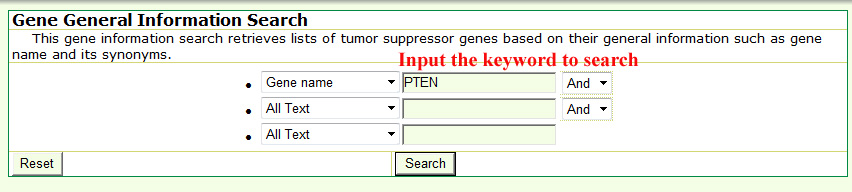
All the tumor suppressor genes and their annotations in our database are searchable. The text search (Query) and sequence-based blast (Blast) are provided. Text search of various annotation in our databaseUsers can search against the TSGene by typing its name, accession IDs and its characteristics, including genomic location, regulatory, interaction partner, mutation, biological pathway, and genetic disease. In total, we provided four different search forms for users, including "Gene General Information Search", "Literature Search", "Mutation Search", and "Other Annotation Search" allow users to access general information, literature-based information, mutation, and other annotation information respectively.
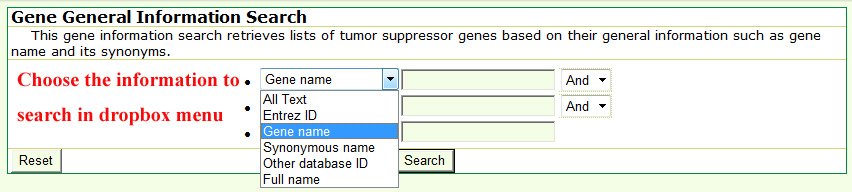
The search is performed by typing keywords into any field separately or into several fields simultaneously in the query forms. Generally, text search information in the each searching form mainly includes three steps. Take the basic information query as an example below select a specific annotation or field from from the dropdown menu in basic gene information and mutation query forms.
Input your interesting keyword.
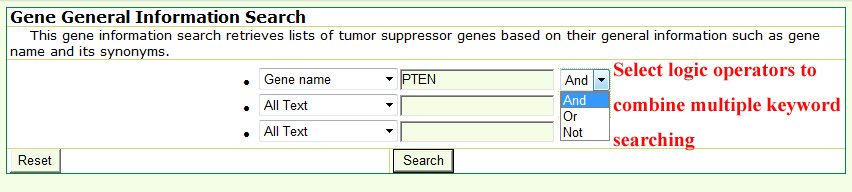
In addition, the basic gene information and mutation query forms support the logical 'And,' 'Or,' and 'Not' operators to combine multiple keywords.
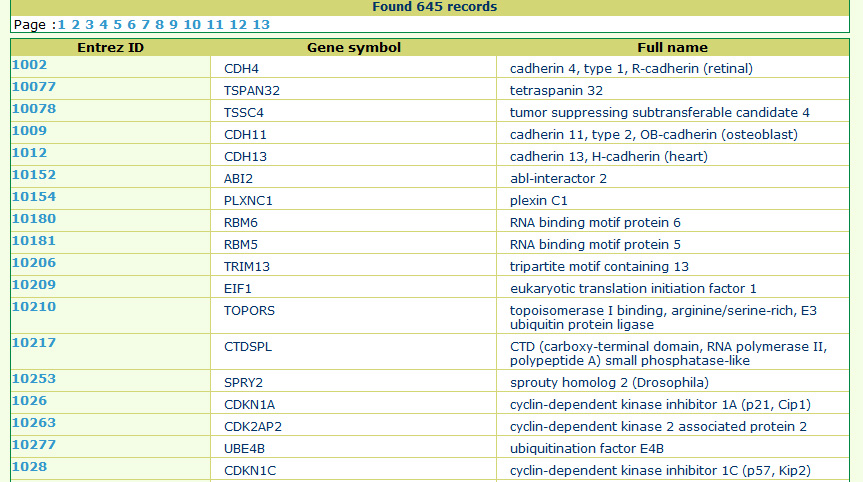
The search result shows the list of matched tumor suppressor genes linked to the detailed gene information page below.
Quick search a list of genes in database:To quickly access the information in the database, a quick search form is provided at the top of each page.
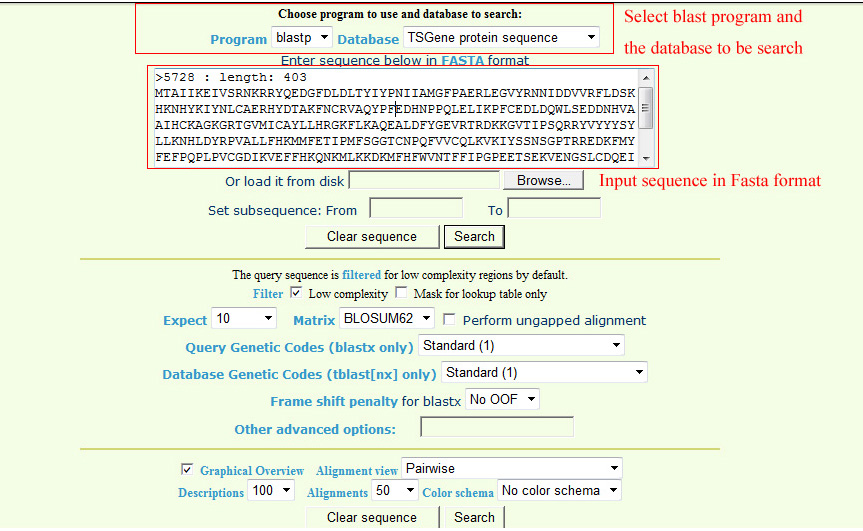
Blast all sequences of genes in our databaseIn the BLAST menu, users can search the TSGene database based on their input sequences. The high similarity tumor suppressor genes with input sequences will be listed in the BLAST result page. In the input page, users can choose various sequence alignment options such as E-value and identity. The matched sequence signatures are visualized on the query sequence with colored bars containing the alignment score. To do a sequence-based search for all the tumor suppressor genes, please access the BLAST page.
The output of BLAST is as below |
Browse database [ top ] |
|
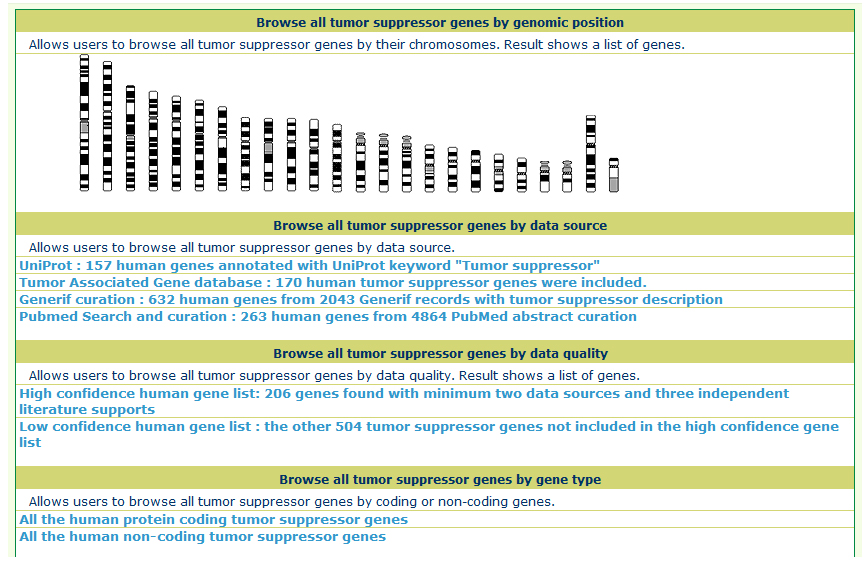
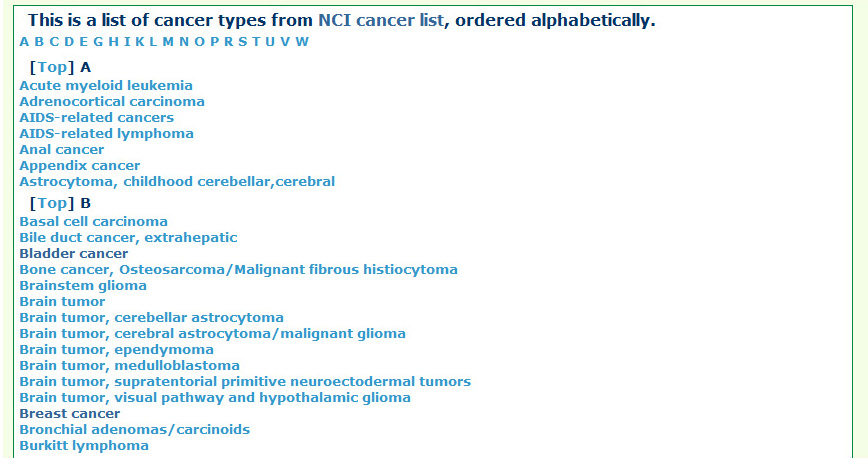
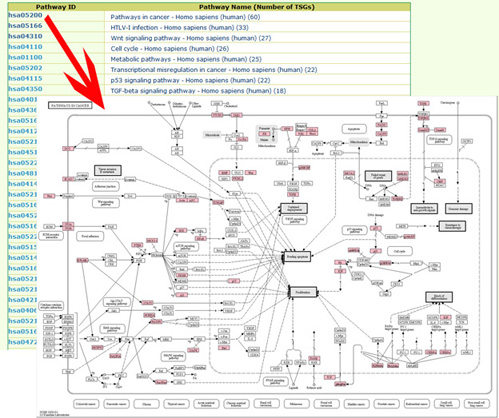
The TSGene database supports browsing tumor suppressor genes using cancer types and KEGG pathway maps. In the cancer type page, users can explore the tumor suppressor genes within specific cancers easily among organized cancer types from NCI. In addition, to help users get a bird's eye view for biological processes of tumor suppressor genes, the marked KEGG maps were provided. Users can browse the tumor suppressor gene of TSGene with their annotated feature list rather than the cancer classification and marked KEGG maps. TSGene also supports annotation-based browsing including chromosome, gene type, data source, data quality. |
Using different data sources, chromosomes, and gene typesFrom the Browser page, users can obtain the tumor suppressor gene lists from different data sources such as the TAG database, UniProt, Generif curation, and PubMed search results. Based on the count of data sources and literature support, we defined a high confidence tumor suppressor gene list with 206 human genes. These genes had at least two data sources and three independent literature supports. In addition, users can also find all the protein coding and recently reported non-coding tumor suppressor genes in the browser page.
By different cancer typeFrom cancer type list users can easily browser all the cancer types according to NCI cancer classifiction system.
By pathway mapsFrom pathway list page users can visulize all the KEGG pathways with any tumor suppressor genes recorded in our database.
|
Data download and feedback [ top ] |
You can freely download our data for academic researchers, but not for profit purposes. Please access Download page.We also hope you can help us to improve our database.If you have any suggestion to add new comment to records in current TSGene or to revise wrong information in current TSGene,please send us email directly. |