Gene Page: CDH5
Summary ?
| GeneID | 1003 |
| Symbol | CDH5 |
| Synonyms | 7B4|CD144 |
| Description | cadherin 5 |
| Reference | MIM:601120|HGNC:HGNC:1764|Ensembl:ENSG00000179776|HPRD:03075|Vega:OTTHUMG00000137495 |
| Gene type | protein-coding |
| Map location | 16q22.1 |
| Pascal p-value | 0.375 |
| Fetal beta | 0.043 |
| eGene | Meta |
| Support | CELL ADHESION AND TRANSSYNAPTIC SIGNALING CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DNM:Fromer_2014 | Whole Exome Sequencing analysis | This study reported a WES study of 623 schizophrenia trios, reporting DNMs using genomic DNA. | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 2 |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| CDH5 | chr16 | 66426075 | A | C | NM_001795 | p.336I>L | missense | Schizophrenia | DNM:Fromer_2014 |
Section II. Transcriptome annotation
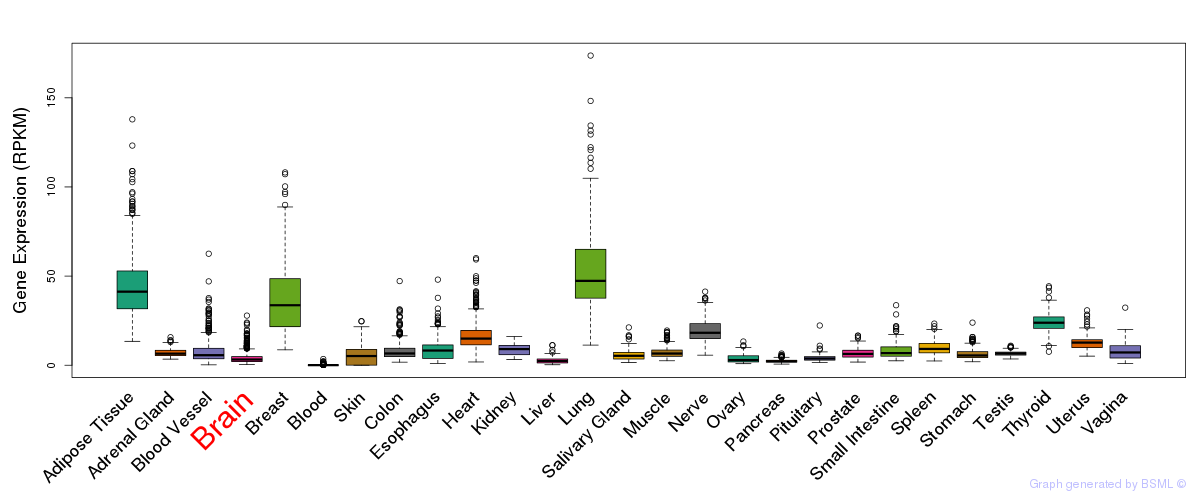
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| PPFIA2 | 0.79 | 0.81 |
| R3HDM1 | 0.78 | 0.84 |
| PVRL3 | 0.78 | 0.84 |
| FGF14 | 0.78 | 0.80 |
| TM6SF1 | 0.77 | 0.81 |
| LRRC4C | 0.76 | 0.81 |
| JAZF1 | 0.76 | 0.81 |
| ZNF540 | 0.76 | 0.78 |
| L3MBTL4 | 0.76 | 0.80 |
| GPR176 | 0.75 | 0.83 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AC098691.2 | -0.54 | -0.59 |
| MT-CO2 | -0.54 | -0.48 |
| FXYD1 | -0.53 | -0.45 |
| AF347015.21 | -0.53 | -0.45 |
| AF347015.33 | -0.52 | -0.44 |
| HIGD1B | -0.52 | -0.48 |
| AC021016.1 | -0.52 | -0.49 |
| AP002478.3 | -0.52 | -0.53 |
| ACSF2 | -0.51 | -0.53 |
| AF347015.8 | -0.51 | -0.45 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005102 | receptor binding | IPI | Neurotransmitter (GO term level: 4) | 12088286 |
| GO:0005509 | calcium ion binding | IEA | - | |
| GO:0005515 | protein binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0001955 | blood vessel maturation | IEA | - | |
| GO:0007155 | cell adhesion | IEA | - | |
| GO:0007156 | homophilic cell adhesion | IEA | - | |
| GO:0007156 | homophilic cell adhesion | TAS | 9219219 | |
| GO:0008285 | negative regulation of cell proliferation | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005923 | tight junction | IEA | Brain (GO term level: 10) | - |
| GO:0005624 | membrane fraction | TAS | 9219219 | |
| GO:0016020 | membrane | IEA | - | |
| GO:0016021 | integral to membrane | IEA | - | |
| GO:0005886 | plasma membrane | TAS | 9219219 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| CSK | MGC117393 | c-src tyrosine kinase | CDH5 (VE-cadherin) interacts with CSK. | BIND | 15861137 |
| CTNNA1 | CAP102 | FLJ36832 | catenin (cadherin-associated protein), alpha 1, 102kDa | - | HPRD,BioGRID | 12003790 |
| CTNNB1 | CTNNB | DKFZp686D02253 | FLJ25606 | FLJ37923 | catenin (cadherin-associated protein), beta 1, 88kDa | - | HPRD | 12413882 |
| CTNNB1 | CTNNB | DKFZp686D02253 | FLJ25606 | FLJ37923 | catenin (cadherin-associated protein), beta 1, 88kDa | Affinity Capture-Western | BioGRID | 9434630 |12003790 |
| CTNND1 | CAS | CTNND | KIAA0384 | P120CAS | P120CTN | p120 | catenin (cadherin-associated protein), delta 1 | - | HPRD,BioGRID | 11855855 |
| DSP | DPI | DPII | desmoplakin | - | HPRD,BioGRID | 9739078 |
| FGA | Fib2 | MGC119422 | MGC119423 | MGC119425 | fibrinogen alpha chain | - | HPRD,BioGRID | 11900554 |
| FGB | MGC104327 | MGC120405 | fibrinogen beta chain | B-beta(15-42) interacts with CDH5 (VE-cadherin). | BIND | 15723073 |
| JUP | ARVD12 | CTNNG | DP3 | DPIII | PDGB | PKGB | junction plakoglobin | - | HPRD | 9739078 |12003790 |
| JUP | ARVD12 | CTNNG | DP3 | DPIII | PDGB | PKGB | junction plakoglobin | Affinity Capture-Western | BioGRID | 9434630 |12003790 |
| KDR | CD309 | FLK1 | VEGFR | VEGFR2 | kinase insert domain receptor (a type III receptor tyrosine kinase) | Affinity Capture-Western | BioGRID | 11950700 |
| KDR | CD309 | FLK1 | VEGFR | VEGFR2 | kinase insert domain receptor (a type III receptor tyrosine kinase) | - | HPRD | 10428027 |
| P2RX4 | P2X4 | P2X4R | purinergic receptor P2X, ligand-gated ion channel, 4 | Affinity Capture-Western | BioGRID | 12088286 |
| P2RX6 | MGC129625 | P2RXL1 | P2X6 | P2XM | purinergic receptor P2X, ligand-gated ion channel, 6 | - | HPRD | 12088286 |
| PKP4 | FLJ31261 | FLJ42243 | p0071 | plakophilin 4 | - | HPRD,BioGRID | 12426320 |
| PKP4 | FLJ31261 | FLJ42243 | p0071 | plakophilin 4 | p0071 interacts with VE-cadherin. | BIND | 12426320 |
| PTPN11 | BPTP3 | CFC | MGC14433 | NS1 | PTP-1D | PTP2C | SH-PTP2 | SH-PTP3 | SHP2 | protein tyrosine phosphatase, non-receptor type 11 | - | HPRD,BioGRID | 10681592 |
| PTPRB | DKFZp686E2262 | DKFZp686H15164 | FLJ44133 | HPTP-BETA | HPTPB | MGC142023 | MGC59935 | PTPB | R-PTP-BETA | VEPTP | protein tyrosine phosphatase, receptor type, B | Affinity Capture-Western Biochemical Activity | BioGRID | 12234928 |
| SHC1 | FLJ26504 | SHC | SHCA | SHC (Src homology 2 domain containing) transforming protein 1 | - | HPRD,BioGRID | 11950700 |
| SMG1 | 61E3.4 | ATX | KIAA0421 | LIP | PI-3-kinase-related kinase SMG-1 | - | HPRD | 10428027 |
| VIM | FLJ36605 | vimentin | - | HPRD,BioGRID | 12003790 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CELL ADHESION MOLECULES CAMS | 134 | 93 | All SZGR 2.0 genes in this pathway |
| KEGG LEUKOCYTE TRANSENDOTHELIAL MIGRATION | 118 | 78 | All SZGR 2.0 genes in this pathway |
| PID VEGFR1 2 PATHWAY | 69 | 57 | All SZGR 2.0 genes in this pathway |
| PID S1P S1P2 PATHWAY | 24 | 19 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CELL COMMUNICATION | 120 | 77 | All SZGR 2.0 genes in this pathway |
| REACTOME ADHERENS JUNCTIONS INTERACTIONS | 27 | 20 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CELL JUNCTION ORGANIZATION | 56 | 31 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL JUNCTION ORGANIZATION | 78 | 43 | All SZGR 2.0 genes in this pathway |
| PICCALUGA ANGIOIMMUNOBLASTIC LYMPHOMA UP | 205 | 140 | All SZGR 2.0 genes in this pathway |
| ROY WOUND BLOOD VESSEL UP | 50 | 30 | All SZGR 2.0 genes in this pathway |
| HORIUCHI WTAP TARGETS DN | 310 | 188 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY SERUM DEPRIVATION DN | 234 | 147 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 3 4WK UP | 214 | 144 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 4 5WK DN | 196 | 131 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL TGFB1 TARGETS DN | 62 | 44 | All SZGR 2.0 genes in this pathway |
| CALVET IRINOTECAN SENSITIVE VS REVERTED DN | 5 | 5 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| RANKIN ANGIOGENIC TARGETS OF VHL HIF2A DN | 8 | 7 | All SZGR 2.0 genes in this pathway |
| NING CHRONIC OBSTRUCTIVE PULMONARY DISEASE UP | 157 | 105 | All SZGR 2.0 genes in this pathway |
| GUO HEX TARGETS DN | 65 | 36 | All SZGR 2.0 genes in this pathway |
| LI WILMS TUMOR VS FETAL KIDNEY 1 UP | 182 | 119 | All SZGR 2.0 genes in this pathway |
| IGLESIAS E2F TARGETS UP | 151 | 103 | All SZGR 2.0 genes in this pathway |
| KUMAR TARGETS OF MLL AF9 FUSION | 405 | 264 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 DN | 830 | 547 | All SZGR 2.0 genes in this pathway |
| YAMAZAKI TCEB3 TARGETS UP | 175 | 116 | All SZGR 2.0 genes in this pathway |
| MONNIER POSTRADIATION TUMOR ESCAPE UP | 393 | 244 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS DN | 543 | 317 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS DN | 593 | 372 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS UP | 601 | 369 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER UP | 973 | 570 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER TUMOR VS NORMAL ADJACENT TISSUE UP | 863 | 514 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL DN | 216 | 143 | All SZGR 2.0 genes in this pathway |
| VART KSHV INFECTION ANGIOGENIC MARKERS DN | 138 | 92 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS DN | 435 | 289 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS UP | 504 | 321 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-101 | 1566 | 1573 | 1A,m8 | hsa-miR-101 | UACAGUACUGUGAUAACUGAAG |
| miR-128 | 715 | 721 | 1A | hsa-miR-128a | UCACAGUGAACCGGUCUCUUUU |
| hsa-miR-128b | UCACAGUGAACCGGUCUCUUUC | ||||
| miR-143 | 1077 | 1083 | 1A | hsa-miR-143brain | UGAGAUGAAGCACUGUAGCUCA |
| miR-144 | 1567 | 1573 | 1A | hsa-miR-144 | UACAGUAUAGAUGAUGUACUAG |
| miR-27 | 715 | 722 | 1A,m8 | hsa-miR-27abrain | UUCACAGUGGCUAAGUUCCGC |
| hsa-miR-27bbrain | UUCACAGUGGCUAAGUUCUGC | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.