Gene Page: CDH13
Summary ?
| GeneID | 1012 |
| Symbol | CDH13 |
| Synonyms | CDHH|P105 |
| Description | cadherin 13 |
| Reference | MIM:601364|HGNC:HGNC:1753|Ensembl:ENSG00000140945|HPRD:03219|Vega:OTTHUMG00000176635 |
| Gene type | protein-coding |
| Map location | 16q23.3 |
| Fetal beta | -1.819 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans Meta |
| Support | CELL ADHESION AND TRANSSYNAPTIC SIGNALING G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWAScat | Genome-wide Association Studies | This data set includes 560 SNPs associated with schizophrenia. A total of 486 genes were mapped to these SNPs within 50kb. | |
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg05949171 | 16 | 82660596 | CDH13 | 4.32E-10 | -0.036 | 7.89E-7 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs12201810 | chr6 | 51409683 | CDH13 | 1012 | 0.14 | trans |
Section II. Transcriptome annotation
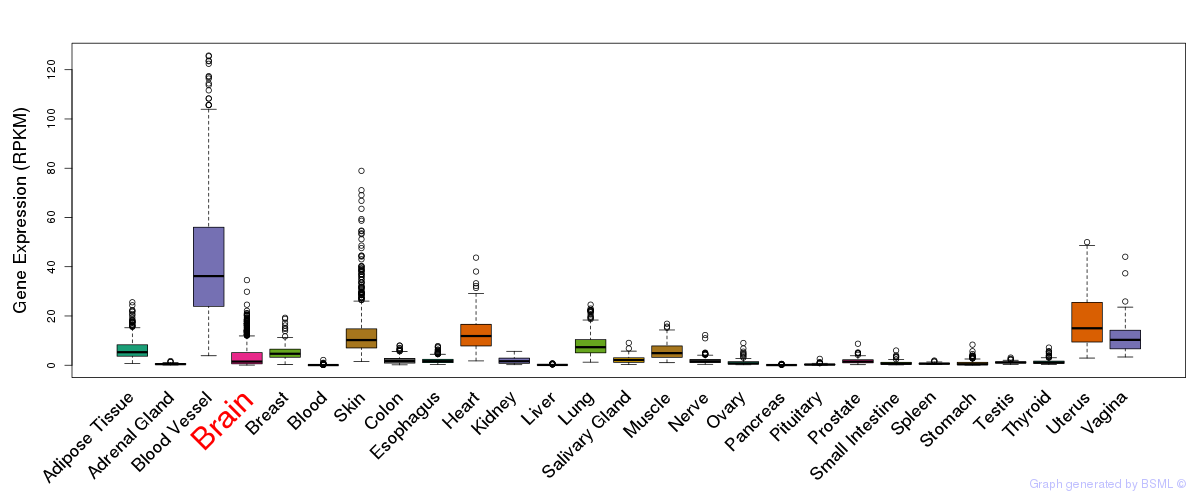
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| NCAPH | 0.98 | 0.66 |
| DTL | 0.97 | 0.69 |
| GTSE1 | 0.97 | 0.70 |
| NUSAP1 | 0.97 | 0.71 |
| TIMELESS | 0.97 | 0.63 |
| KIF2C | 0.97 | 0.70 |
| CDC45L | 0.97 | 0.68 |
| ORC1L | 0.97 | 0.63 |
| MCM10 | 0.97 | 0.71 |
| TACC3 | 0.97 | 0.66 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| FBXO2 | -0.40 | -0.55 |
| C5orf53 | -0.40 | -0.57 |
| ASPHD1 | -0.39 | -0.54 |
| HLA-F | -0.38 | -0.50 |
| SLC9A3R2 | -0.38 | -0.39 |
| CCNI2 | -0.38 | -0.59 |
| ADAP1 | -0.38 | -0.43 |
| LHPP | -0.37 | -0.48 |
| PTH1R | -0.37 | -0.45 |
| CA4 | -0.37 | -0.57 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005509 | calcium ion binding | IEA | - | |
| GO:0042803 | protein homodimerization activity | IEA | - | |
| GO:0030169 | low-density lipoprotein binding | IDA | 16013438 | |
| GO:0045296 | cadherin binding | IDA | 10601632 | |
| GO:0055100 | adiponectin binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0002040 | sprouting angiogenesis | IDA | 16873731 | |
| GO:0001938 | positive regulation of endothelial cell proliferation | IMP | 15364621 | |
| GO:0001954 | positive regulation of cell-matrix adhesion | IMP | 17573778 | |
| GO:0030100 | regulation of endocytosis | IMP | 17573778 | |
| GO:0007162 | negative regulation of cell adhesion | IDA | 14729458 | |
| GO:0007156 | homophilic cell adhesion | IEA | - | |
| GO:0007266 | Rho protein signal transduction | IMP | 15703273 | |
| GO:0008285 | negative regulation of cell proliferation | IDA | 10737605 | |
| GO:0016601 | Rac protein signal transduction | IMP | 15703273 | |
| GO:0016339 | calcium-dependent cell-cell adhesion | IDA | 10601632 | |
| GO:0050927 | positive regulation of positive chemotaxis | IDA | 16013438 | |
| GO:0042058 | regulation of epidermal growth factor receptor signaling pathway | IMP | 17573778 | |
| GO:0030335 | positive regulation of cell migration | IDA | 14729458 | |
| GO:0030032 | lamellipodium biogenesis | IDA | 15703273 | |
| GO:0045885 | positive regulation of survival gene product expression | IMP | 16099944 | |
| GO:0050850 | positive regulation of calcium-mediated signaling | IDA | 16013438 | |
| GO:0043542 | endothelial cell migration | IDA | 14729458 | |
| GO:0043616 | keratinocyte proliferation | IDA | 15816843 | |
| GO:0048661 | positive regulation of smooth muscle cell proliferation | IMP | 15364621 | |
| GO:0055096 | low density lipoprotein mediated signaling | IDA | 16013438 | |
| GO:0051668 | localization within membrane | IMP | 17573778 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0043005 | neuron projection | IDA | neuron, axon, neurite, dendrite (GO term level: 5) | 10737605 |
| GO:0005615 | extracellular space | IDA | 16873731 | |
| GO:0005737 | cytoplasm | IDA | 10737605 | |
| GO:0009897 | external side of plasma membrane | IEA | - | |
| GO:0005901 | caveola | IDA | 9650591 | |
| GO:0005886 | plasma membrane | IEA | - | |
| GO:0031225 | anchored to membrane | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| REACTOME CELL CELL COMMUNICATION | 120 | 77 | All SZGR 2.0 genes in this pathway |
| REACTOME ADHERENS JUNCTIONS INTERACTIONS | 27 | 20 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CELL JUNCTION ORGANIZATION | 56 | 31 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL JUNCTION ORGANIZATION | 78 | 43 | All SZGR 2.0 genes in this pathway |
| DAVICIONI MOLECULAR ARMS VS ERMS UP | 332 | 228 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS BASAL DN | 455 | 304 | All SZGR 2.0 genes in this pathway |
| HORIUCHI WTAP TARGETS UP | 306 | 188 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 TARGETS DN | 260 | 143 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC UP | 722 | 443 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE DN | 485 | 334 | All SZGR 2.0 genes in this pathway |
| CHIN BREAST CANCER COPY NUMBER DN | 16 | 10 | All SZGR 2.0 genes in this pathway |
| MAHADEVAN GIST MORPHOLOGICAL SWITCH | 15 | 11 | All SZGR 2.0 genes in this pathway |
| SILIGAN BOUND BY EWS FLT1 FUSION | 47 | 34 | All SZGR 2.0 genes in this pathway |
| DAIRKEE TERT TARGETS UP | 380 | 213 | All SZGR 2.0 genes in this pathway |
| HATADA METHYLATED IN LUNG CANCER UP | 390 | 236 | All SZGR 2.0 genes in this pathway |
| SCHLESINGER METHYLATED DE NOVO IN CANCER | 88 | 64 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 48HR UP | 487 | 286 | All SZGR 2.0 genes in this pathway |
| BENPORATH SUZ12 TARGETS | 1038 | 678 | All SZGR 2.0 genes in this pathway |
| CHEN NEUROBLASTOMA COPY NUMBER GAINS | 50 | 35 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| TRAYNOR RETT SYNDROM DN | 19 | 14 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 24HR DN | 148 | 102 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 14HR DN | 298 | 200 | All SZGR 2.0 genes in this pathway |
| KAYO AGING MUSCLE DN | 123 | 76 | All SZGR 2.0 genes in this pathway |
| KAAB HEART ATRIUM VS VENTRICLE DN | 261 | 183 | All SZGR 2.0 genes in this pathway |
| MCCLUNG CREB1 TARGETS UP | 100 | 72 | All SZGR 2.0 genes in this pathway |
| MCDOWELL ACUTE LUNG INJURY DN | 48 | 33 | All SZGR 2.0 genes in this pathway |
| MARCHINI TRABECTEDIN RESISTANCE DN | 49 | 34 | All SZGR 2.0 genes in this pathway |
| ACEVEDO METHYLATED IN LIVER CANCER DN | 940 | 425 | All SZGR 2.0 genes in this pathway |
| HOQUE METHYLATED IN CANCER | 56 | 45 | All SZGR 2.0 genes in this pathway |
| BONOME OVARIAN CANCER SURVIVAL SUBOPTIMAL DEBULKING | 510 | 309 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS 36HR UP | 221 | 150 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS 60HR UP | 293 | 203 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS UP | 108 | 78 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN PARTIALLY REPROGRAMMED TO PLURIPOTENCY | 10 | 6 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3 | 1069 | 729 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER LATE RECURRENCE UP | 62 | 42 | All SZGR 2.0 genes in this pathway |
| LI PROSTATE CANCER EPIGENETIC | 30 | 22 | All SZGR 2.0 genes in this pathway |
| NAKAMURA ADIPOGENESIS LATE UP | 104 | 67 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 30MIN DN | 150 | 99 | All SZGR 2.0 genes in this pathway |
| AZARE NEOPLASTIC TRANSFORMATION BY STAT3 UP | 121 | 70 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS UP | 745 | 475 | All SZGR 2.0 genes in this pathway |
| KOINUMA TARGETS OF SMAD2 OR SMAD3 | 824 | 528 | All SZGR 2.0 genes in this pathway |
| IKEDA MIR30 TARGETS DN | 27 | 24 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG BOUND 8D | 658 | 397 | All SZGR 2.0 genes in this pathway |
| PEDRIOLI MIR31 TARGETS DN | 418 | 245 | All SZGR 2.0 genes in this pathway |
| ROESSLER LIVER CANCER METASTASIS UP | 107 | 72 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY STEM CELL UP | 489 | 314 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-181 | 224 | 230 | m8 | hsa-miR-181abrain | AACAUUCAACGCUGUCGGUGAGU |
| hsa-miR-181bSZ | AACAUUCAUUGCUGUCGGUGGG | ||||
| hsa-miR-181cbrain | AACAUUCAACCUGUCGGUGAGU | ||||
| hsa-miR-181dbrain | AACAUUCAUUGUUGUCGGUGGGUU | ||||
| miR-30-5p | 83 | 89 | 1A | hsa-miR-30a-5p | UGUAAACAUCCUCGACUGGAAG |
| hsa-miR-30cbrain | UGUAAACAUCCUACACUCUCAGC | ||||
| hsa-miR-30dSZ | UGUAAACAUCCCCGACUGGAAG | ||||
| hsa-miR-30bSZ | UGUAAACAUCCUACACUCAGCU | ||||
| hsa-miR-30e-5p | UGUAAACAUCCUUGACUGGA | ||||
| miR-370 | 265 | 271 | 1A | hsa-miR-370brain | GCCUGCUGGGGUGGAACCUGG |
| miR-493-5p | 873 | 880 | 1A,m8 | hsa-miR-493-5p | UUGUACAUGGUAGGCUUUCAUU |
| miR-495 | 993 | 999 | m8 | hsa-miR-495brain | AAACAAACAUGGUGCACUUCUUU |
| miR-543 | 225 | 231 | m8 | hsa-miR-543 | AAACAUUCGCGGUGCACUUCU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.