Gene Page: CLN3
Summary ?
| GeneID | 1201 |
| Symbol | CLN3 |
| Synonyms | BTS|JNCL |
| Description | ceroid-lipofuscinosis, neuronal 3 |
| Reference | MIM:607042|HGNC:HGNC:2074|Ensembl:ENSG00000188603|Ensembl:ENSG00000261832|HPRD:08450|Vega:OTTHUMG00000097024Vega:OTTHUMG00000175737 |
| Gene type | protein-coding |
| Map location | 16p12.1 |
| Pascal p-value | 0.324 |
| Sherlock p-value | 0.516 |
| Fetal beta | 0.654 |
| DMG | 2 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 2 |
| DMG:vanEijk_2014 | Genome-wide DNA methylation analysis | This dataset includes 432 differentially methylated CpG sites corresponding to 391 unique transcripts between schizophrenia patients (n=260) and unaffected controls (n=250). | 2 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| GSMA_IIE | Genome scan meta-analysis (European-ancestry samples) | Psr: 0.01775 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 5 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg09112682 | 16 | 28503005 | CLN3 | 3.43E-9 | -0.009 | 2.26E-6 | DMG:Jaffe_2016 |
| cg15812873 | 16 | 28635015 | CLN3 | 1.928E-4 | 5.337 | DMG:vanEijk_2014 |
Section II. Transcriptome annotation
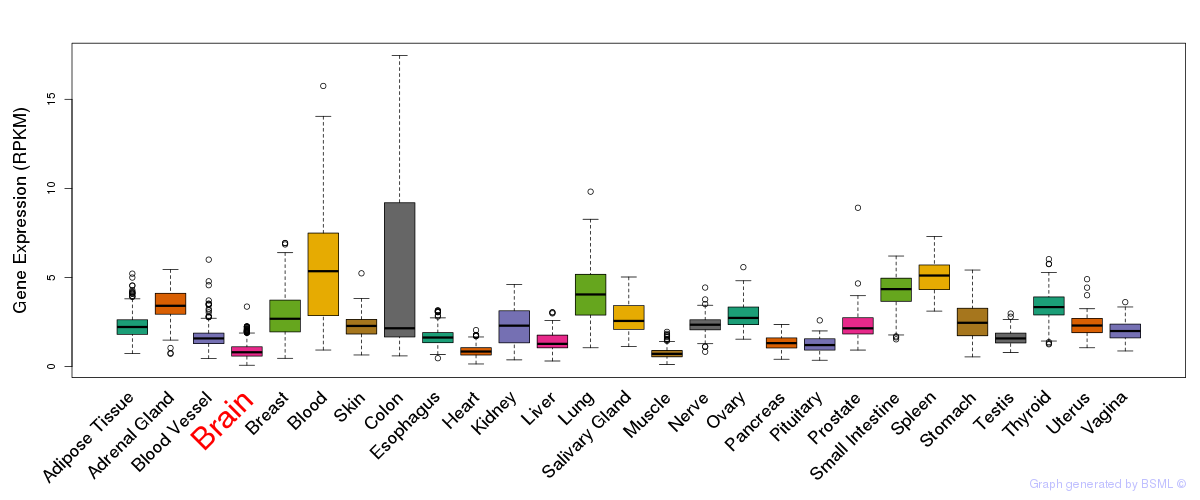
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0048306 | calcium-dependent protein binding | IEA | - | |
| GO:0051082 | unfolded protein binding | TAS | 8980123 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0042133 | neurotransmitter metabolic process | IEA | neuron, axon, Synap, Neurotransmitter (GO term level: 8) | - |
| GO:0043524 | negative regulation of neuron apoptosis | IEA | neuron (GO term level: 9) | - |
| GO:0035235 | ionotropic glutamate receptor signaling pathway | IEA | glutamate (GO term level: 8) | - |
| GO:0000046 | autophagic vacuole fusion | IEA | - | |
| GO:0001575 | globoside metabolic process | IMP | 15240864 | |
| GO:0001508 | regulation of action potential | IEA | - | |
| GO:0042987 | amyloid precursor protein catabolic process | IDA | 10924275 | |
| GO:0006457 | protein folding | TAS | 8980123 | |
| GO:0016044 | membrane organization | IEA | - | |
| GO:0006520 | amino acid metabolic process | IEA | - | |
| GO:0007042 | lysosomal lumen acidification | IMP | 11722572 | |
| GO:0008306 | associative learning | IEA | - | |
| GO:0015809 | arginine transport | IDA | 16251196 | |
| GO:0006684 | sphingomyelin metabolic process | IMP | 15240864 | |
| GO:0006681 | galactosylceramide metabolic process | IMP | 15240864 | |
| GO:0006678 | glucosylceramide metabolic process | IMP | 15240864 | |
| GO:0006898 | receptor-mediated endocytosis | IMP | 15471887 | |
| GO:0006865 | amino acid transport | IEA | - | |
| GO:0016242 | negative regulation of macroautophagy | IEA | - | |
| GO:0016485 | protein processing | IEA | - | |
| GO:0050885 | neuromuscular process controlling balance | IEA | - | |
| GO:0030163 | protein catabolic process | NAS | 10740217 | |
| GO:0045861 | negative regulation of proteolysis | IEA | - | |
| GO:0051453 | regulation of intracellular pH | IDA | 10924275 | |
| GO:0051480 | cytosolic calcium ion homeostasis | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0008021 | synaptic vesicle | IDA | Synap, Neurotransmitter (GO term level: 12) | 10332042 |11590129 |
| GO:0043005 | neuron projection | IDA | neuron, axon, neurite, dendrite (GO term level: 5) | 10332042 |
| GO:0000139 | Golgi membrane | IDA | 9949212 |10191111 | |
| GO:0005794 | Golgi apparatus | IDA | 15240864 | |
| GO:0005795 | Golgi stack | IDA | 15240864 | |
| GO:0005802 | trans-Golgi network | IDA | 15240864 | |
| GO:0005624 | membrane fraction | IDA | 12134079 | |
| GO:0005634 | nucleus | IDA | 10191116 | |
| GO:0005737 | cytoplasm | IDA | 10191116 | |
| GO:0005739 | mitochondrion | TAS | 8980123 | |
| GO:0005765 | lysosomal membrane | IEA | - | |
| GO:0005769 | early endosome | IDA | 15240864 | |
| GO:0005770 | late endosome | IEA | - | |
| GO:0005776 | autophagic vacuole | IEA | - | |
| GO:0005783 | endoplasmic reticulum | IDA | 10191111 | |
| GO:0016020 | membrane | IEA | - | |
| GO:0016021 | integral to membrane | IDA | 10191112 | |
| GO:0005901 | caveola | IDA | 15240864 | |
| GO:0005886 | plasma membrane | IDA | 10191116 |15240864 | |
| GO:0030176 | integral to endoplasmic reticulum membrane | IDA | 12706816 | |
| GO:0044433 | cytoplasmic vesicle part | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG LYSOSOME | 121 | 83 | All SZGR 2.0 genes in this pathway |
| PRAMOONJAGO SOX4 TARGETS DN | 51 | 35 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS MESENCHYMAL UP | 450 | 256 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 1 UP | 380 | 236 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 2 UP | 418 | 263 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 1 DN | 378 | 231 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER APOCRINE VS BASAL | 330 | 217 | All SZGR 2.0 genes in this pathway |
| PATIL LIVER CANCER | 747 | 453 | All SZGR 2.0 genes in this pathway |
| BALLIF DEVELOPMENTAL DISABILITY P16 P12 DELETION | 13 | 8 | All SZGR 2.0 genes in this pathway |
| WEI MYCN TARGETS WITH E BOX | 795 | 478 | All SZGR 2.0 genes in this pathway |
| RICKMAN TUMOR DIFFERENTIATED WELL VS MODERATELY UP | 109 | 69 | All SZGR 2.0 genes in this pathway |
| BENPORATH NANOG TARGETS | 988 | 594 | All SZGR 2.0 genes in this pathway |
| BENPORATH SOX2 TARGETS | 734 | 436 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS | 775 | 494 | All SZGR 2.0 genes in this pathway |
| PENG GLUTAMINE DEPRIVATION UP | 38 | 25 | All SZGR 2.0 genes in this pathway |
| NEMETH INFLAMMATORY RESPONSE LPS UP | 88 | 64 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 48HR UP | 180 | 125 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER TUMOR VS NORMAL ADJACENT TISSUE UP | 863 | 514 | All SZGR 2.0 genes in this pathway |
| HUANG DASATINIB RESISTANCE DN | 69 | 44 | All SZGR 2.0 genes in this pathway |
| MOOTHA HUMAN MITODB 6 2002 | 429 | 260 | All SZGR 2.0 genes in this pathway |
| MOOTHA MITOCHONDRIA | 447 | 277 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE LATE | 1137 | 655 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS UP | 745 | 475 | All SZGR 2.0 genes in this pathway |
| KOHOUTEK CCNT1 TARGETS | 50 | 26 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG RXRA BOUND 8D | 882 | 506 | All SZGR 2.0 genes in this pathway |
| FORTSCHEGGER PHF8 TARGETS UP | 279 | 155 | All SZGR 2.0 genes in this pathway |
| ZWANG EGF INTERVAL DN | 214 | 124 | All SZGR 2.0 genes in this pathway |