Gene Page: DRD2
Summary ?
| GeneID | 1813 |
| Symbol | DRD2 |
| Synonyms | D2DR|D2R |
| Description | dopamine receptor D2 |
| Reference | MIM:126450|HGNC:HGNC:3023|Ensembl:ENSG00000149295|HPRD:00539|Vega:OTTHUMG00000167717 |
| Gene type | protein-coding |
| Map location | 11q23 |
| Pascal p-value | 1.601E-7 |
| Fetal beta | -0.638 |
| eGene | Myers' cis & trans |
| Support | GPCR SIGNALLING Potential synaptic genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| ADT:Sun_2012 | Systematic Investigation of Antipsychotic Drugs and Their Targets | A total of 382 drug-target associations involving 43 antipsychotic drugs and 49 target genes. | |
| CV:GWAScat | Genome-wide Association Studies | This data set includes 560 SNPs associated with schizophrenia. A total of 486 genes were mapped to these SNPs within 50kb. | |
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGC128 | Genome-wide Association Study | A multi-stage schizophrenia GWAS of up to 36,989 cases and 113,075 controls. Reported by the Schizophrenia Working Group of PGC. 128 independent associations spanning 108 loci | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Association | A combined odds ratio method (Sun et al. 2008), association studies | 3 | Link to SZGene |
| GSMA_I | Genome scan meta-analysis | Psr: 0.006 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenics,schizotypy,schizophrenias,schizotypal | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 16 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 31 |
Section I. Genetics and epigenetics annotation
 CV:PGC128
CV:PGC128
| SNP ID | Chromosome | Position | Allele | P | Function | Gene | Up/Down Distance |
|---|---|---|---|---|---|---|---|
| rs2514218 | chr11 | 113392994 | TC | 4.091E-10 | intergenic | DRD2,TMPRSS5 | dist=46993;dist=165274 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs2190759 | chr17 | 55794698 | DRD2 | 1813 | 0.08 | trans |
Section II. Transcriptome annotation
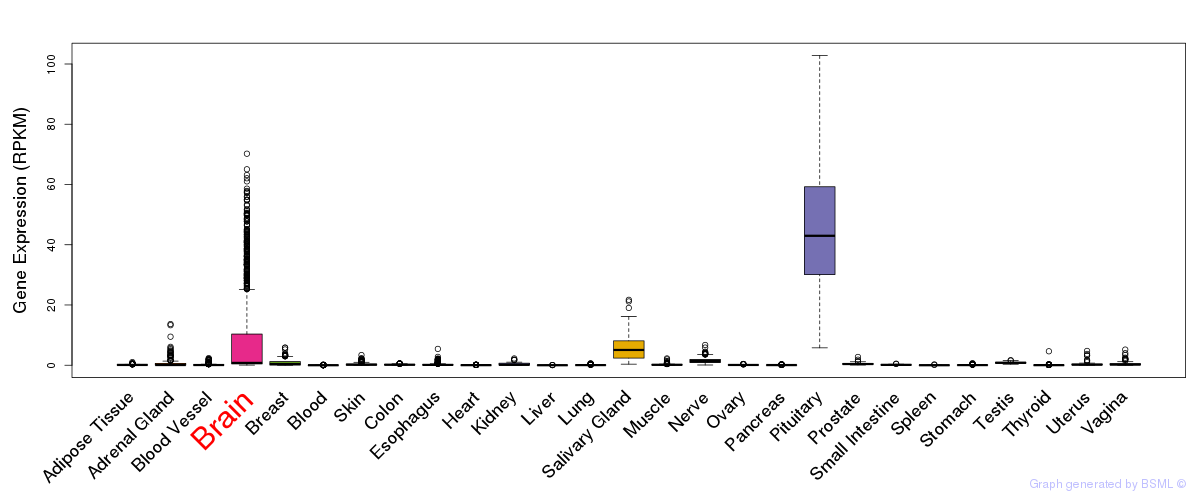
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004952 | dopamine receptor activity | IEA | Neurotransmitter, dopamine (GO term level: 8) | - |
| GO:0001670 | dopamine D2 receptor activity | IDA | dopamine (GO term level: 10) | 8301582 |
| GO:0004872 | receptor activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0048755 | branching morphogenesis of a nerve | ISS | neuron (GO term level: 6) | - |
| GO:0007416 | synaptogenesis | ISS | Synap (GO term level: 6) | - |
| GO:0007409 | axonogenesis | ISS | neuron, axon, neurite (GO term level: 12) | - |
| GO:0060160 | negative regulation of dopamine receptor signaling pathway | ISS | dopamine (GO term level: 10) | - |
| GO:0060158 | dopamine receptor, phospholipase C activating pathway | IGI | dopamine (GO term level: 10) | 17194762 |
| GO:0051586 | positive regulation of dopamine uptake | ISS | dopamine (GO term level: 12) | - |
| GO:0051967 | negative regulation of synaptic transmission, glutamatergic | ISS | neuron, glutamate, Synap, Neurotransmitter (GO term level: 10) | - |
| GO:0007270 | nerve-nerve synaptic transmission | ISS | neuron, Synap (GO term level: 7) | - |
| GO:0048169 | regulation of long-term neuronal synaptic plasticity | ISS | neuron, Synap (GO term level: 10) | - |
| GO:0032228 | regulation of synaptic transmission, GABAergic | ISS | neuron, GABA, Synap, Neurotransmitter (GO term level: 9) | - |
| GO:0021853 | cerebral cortex GABAergic interneuron migration | ISS | neuron, GABA (GO term level: 14) | - |
| GO:0042417 | dopamine metabolic process | IC | Neurotransmitter, dopamine (GO term level: 8) | 8301582 |
| GO:0001659 | temperature homeostasis | ISS | - | |
| GO:0002052 | positive regulation of neuroblast proliferation | ISS | - | |
| GO:0002027 | regulation of heart rate | ISS | - | |
| GO:0002028 | regulation of sodium ion transport | ISS | - | |
| GO:0001975 | response to amphetamine | ISS | - | |
| GO:0001976 | regulation of systemic arterial blood pressure by neurological process | ISS | - | |
| GO:0007186 | G-protein coupled receptor protein signaling pathway | IEA | - | |
| GO:0007165 | signal transduction | IEA | - | |
| GO:0008285 | negative regulation of cell proliferation | ISS | - | |
| GO:0008306 | associative learning | ISS | - | |
| GO:0008542 | visual learning | ISS | - | |
| GO:0009416 | response to light stimulus | ISS | - | |
| GO:0007628 | adult walking behavior | ISS | - | |
| GO:0007626 | locomotory behavior | ISS | - | |
| GO:0008104 | protein localization | ISS | - | |
| GO:0007608 | sensory perception of smell | ISS | - | |
| GO:0042493 | response to drug | ISS | - | |
| GO:0021984 | adenohypophysis development | ISS | - | |
| GO:0030432 | peristalsis | ISS | - | |
| GO:0051209 | release of sequestered calcium ion into cytosol | ISS | - | |
| GO:0030814 | regulation of cAMP metabolic process | IDA | 8666994 | |
| GO:0030146 | diuresis | ISS | - | |
| GO:0030147 | natriuresis | ISS | - | |
| GO:0032922 | circadian regulation of gene expression | ISS | - | |
| GO:0030384 | phosphoinositide metabolic process | ISS | - | |
| GO:0030336 | negative regulation of cell migration | ISS | - | |
| GO:0043278 | response to morphine | ISS | - | |
| GO:0043266 | regulation of potassium ion transport | ISS | - | |
| GO:0048148 | behavioral response to cocaine | ISS | - | |
| GO:0048149 | behavioral response to ethanol | ISS | - | |
| GO:0050709 | negative regulation of protein secretion | IDA | 16839358 | |
| GO:0050482 | arachidonic acid secretion | IDA | 8301582 | |
| GO:0060124 | positive regulation of growth hormone secretion | ISS | - | |
| GO:0045776 | negative regulation of blood pressure | ISS | - | |
| GO:0051898 | negative regulation of protein kinase B signaling cascade | ISS | - | |
| GO:0060134 | prepulse inhibition | ISS | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0030425 | dendrite | ISS | neuron, axon, dendrite (GO term level: 6) | - |
| GO:0030424 | axon | ISS | neuron, axon, Neurotransmitter (GO term level: 6) | - |
| GO:0016021 | integral to membrane | IEA | - | |
| GO:0005886 | plasma membrane | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ADORA2A | ADORA2 | RDC8 | hA2aR | adenosine A2a receptor | - | HPRD,BioGRID | 12804599 |
| CLIC6 | CLIC1L | chloride intracellular channel 6 | - | HPRD,BioGRID | 14499480 |
| DRD2 | D2DR | D2R | dopamine receptor D2 | D2S receptors form homodimers through Cys126 in the fourth transmembrane segment. | BIND | 12496294 |
| EPB41 | 4.1R | EL1 | HE | erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked) | - | HPRD | 12181426 |
| EPB41L1 | 4.1N | DKFZp686H17242 | KIAA0338 | MGC11072 | erythrocyte membrane protein band 4.1-like 1 | - | HPRD,BioGRID | 12181426 |
| FLNA | ABP-280 | ABPX | DKFZp434P031 | FLN | FLN1 | FMD | MNS | NHBP | OPD | OPD1 | OPD2 | filamin A, alpha (actin binding protein 280) | Affinity Capture-Western | BioGRID | 12181426 |
| FLNA | ABP-280 | ABPX | DKFZp434P031 | FLN | FLN1 | FMD | MNS | NHBP | OPD | OPD1 | OPD2 | filamin A, alpha (actin binding protein 280) | Dopamine receptor D2 interacts with filamin A. This interaction was modeled on a demonstrated interaction between human FLN-A and D2 from unspecified source. | BIND | 11320256 |
| FLNA | ABP-280 | ABPX | DKFZp434P031 | FLN | FLN1 | FMD | MNS | NHBP | OPD | OPD1 | OPD2 | filamin A, alpha (actin binding protein 280) | - | HPRD | 11911837 |
| FREQ | DKFZp761L1223 | FLUP | NCS-1 | NCS1 | frequenin homolog (Drosophila) | - | HPRD,BioGRID | 12351722 |
| FREQ | DKFZp761L1223 | FLUP | NCS-1 | NCS1 | frequenin homolog (Drosophila) | The D2 dopamine receptor interacts with NCS-1. | BIND | 12351722 |
| GNAI2 | GIP | GNAI2B | H_LUCA15.1 | H_LUCA16.1 | guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2 | - | HPRD | 10978845 |
| GNAI3 | 87U6 | FLJ26559 | guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 3 | - | HPRD | 10978845 |
| GNAZ | - | guanine nucleotide binding protein (G protein), alpha z polypeptide | - | HPRD | 9166747 |
| KCNJ3 | GIRK1 | KGA | KIR3.1 | potassium inwardly-rectifying channel, subfamily J, member 3 | Affinity Capture-Western | BioGRID | 12297500 |
| KCNJ5 | CIR | GIRK4 | KATP1 | KIR3.4 | potassium inwardly-rectifying channel, subfamily J, member 5 | Affinity Capture-Western | BioGRID | 12297500 |
| KCNJ6 | BIR1 | GIRK2 | KATP2 | KCNJ7 | KIR3.2 | MGC126596 | hiGIRK2 | potassium inwardly-rectifying channel, subfamily J, member 6 | - | HPRD,BioGRID | 12297500 |
| KCNJ9 | GIRK3 | KIR3.3 | potassium inwardly-rectifying channel, subfamily J, member 9 | - | HPRD | 12297500 |
| PPP1R9B | FLJ30345 | PPP1R6 | PPP1R9 | SPINO | Spn | protein phosphatase 1, regulatory (inhibitor) subunit 9B | - | HPRD,BioGRID | 10391935 |
| SSTR5 | - | somatostatin receptor 5 | - | HPRD | 10753124 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG NEUROACTIVE LIGAND RECEPTOR INTERACTION | 272 | 195 | All SZGR 2.0 genes in this pathway |
| KEGG GAP JUNCTION | 90 | 68 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CK1 PATHWAY | 17 | 17 | All SZGR 2.0 genes in this pathway |
| ST G ALPHA I PATHWAY | 35 | 29 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY GPCR | 920 | 449 | All SZGR 2.0 genes in this pathway |
| REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | 305 | 177 | All SZGR 2.0 genes in this pathway |
| REACTOME AMINE LIGAND BINDING RECEPTORS | 38 | 33 | All SZGR 2.0 genes in this pathway |
| REACTOME GPCR DOWNSTREAM SIGNALING | 805 | 368 | All SZGR 2.0 genes in this pathway |
| REACTOME G ALPHA I SIGNALLING EVENTS | 195 | 114 | All SZGR 2.0 genes in this pathway |
| REACTOME GPCR LIGAND BINDING | 408 | 246 | All SZGR 2.0 genes in this pathway |
| PARENT MTOR SIGNALING UP | 567 | 375 | All SZGR 2.0 genes in this pathway |
| DEURIG T CELL PROLYMPHOCYTIC LEUKEMIA UP | 368 | 234 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS D UP | 38 | 23 | All SZGR 2.0 genes in this pathway |
| XU HGF TARGETS REPRESSED BY AKT1 DN | 95 | 58 | All SZGR 2.0 genes in this pathway |
| MOHANKUMAR TLX1 TARGETS DN | 193 | 112 | All SZGR 2.0 genes in this pathway |
| GRUETZMANN PANCREATIC CANCER DN | 203 | 134 | All SZGR 2.0 genes in this pathway |
| JAZAG TGFB1 SIGNALING VIA SMAD4 DN | 66 | 38 | All SZGR 2.0 genes in this pathway |
| KIM MYCN AMPLIFICATION TARGETS UP | 92 | 64 | All SZGR 2.0 genes in this pathway |
| SHEN SMARCA2 TARGETS DN | 357 | 212 | All SZGR 2.0 genes in this pathway |
| LEE CALORIE RESTRICTION NEOCORTEX UP | 83 | 66 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| KAYO CALORIE RESTRICTION MUSCLE UP | 95 | 64 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3K27ME3 | 79 | 59 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3 | 1069 | 729 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN NPC HCP WITH H3K27ME3 | 341 | 243 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MEF HCP WITH H3K27ME3 | 590 | 403 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE UP | 857 | 456 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-141/200a | 128 | 134 | m8 | hsa-miR-141 | UAACACUGUCUGGUAAAGAUGG |
| hsa-miR-200a | UAACACUGUCUGGUAACGAUGU | ||||
| miR-214 | 978 | 984 | 1A | hsa-miR-214brain | ACAGCAGGCACAGACAGGCAG |
| miR-326 | 527 | 534 | 1A,m8 | hsa-miR-326 | CCUCUGGGCCCUUCCUCCAG |
| miR-9 | 289 | 296 | 1A,m8 | hsa-miR-9SZ | UCUUUGGUUAUCUAGCUGUAUGA |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.