Gene Page: DUSP2
Summary ?
| GeneID | 1844 |
| Symbol | DUSP2 |
| Synonyms | PAC-1|PAC1 |
| Description | dual specificity phosphatase 2 |
| Reference | MIM:603068|HGNC:HGNC:3068|Ensembl:ENSG00000158050|HPRD:04348|Vega:OTTHUMG00000130456 |
| Gene type | protein-coding |
| Map location | 2q11 |
| Pascal p-value | 0.002 |
| Fetal beta | -1.465 |
| eGene | Caudate basal ganglia Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.0004 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs725667 | chr1 | 68488649 | DUSP2 | 1844 | 0.14 | trans | ||
| rs11807500 | chr1 | 68503525 | DUSP2 | 1844 | 0.01 | trans | ||
| rs11808365 | chr1 | 68507122 | DUSP2 | 1844 | 0.14 | trans | ||
| rs12086539 | chr1 | 68507537 | DUSP2 | 1844 | 0.13 | trans | ||
| rs2566762 | chr1 | 68582456 | DUSP2 | 1844 | 0.13 | trans | ||
| rs17100274 | chr1 | 77774412 | DUSP2 | 1844 | 0.18 | trans | ||
| rs6666937 | chr1 | 170286842 | DUSP2 | 1844 | 0.13 | trans | ||
| rs16843112 | chr1 | 198420657 | DUSP2 | 1844 | 0.01 | trans | ||
| rs16843346 | chr1 | 198543026 | DUSP2 | 1844 | 0.08 | trans | ||
| rs16843424 | chr1 | 198572758 | DUSP2 | 1844 | 0.1 | trans | ||
| rs16843670 | chr1 | 198650005 | DUSP2 | 1844 | 0.01 | trans | ||
| rs10925947 | chr1 | 235701552 | DUSP2 | 1844 | 0.06 | trans | ||
| rs636167 | chr1 | 237402264 | DUSP2 | 1844 | 0 | trans | ||
| rs652792 | chr1 | 237403609 | DUSP2 | 1844 | 0.04 | trans | ||
| rs17490288 | chr2 | 36902427 | DUSP2 | 1844 | 0.1 | trans | ||
| rs12465555 | chr2 | 44118188 | DUSP2 | 1844 | 0.07 | trans | ||
| rs17024035 | chr2 | 100968559 | DUSP2 | 1844 | 0.18 | trans | ||
| rs7574684 | chr2 | 100992911 | DUSP2 | 1844 | 0.18 | trans | ||
| rs17024129 | chr2 | 100995144 | DUSP2 | 1844 | 0.18 | trans | ||
| rs7585898 | chr2 | 100998292 | DUSP2 | 1844 | 0.18 | trans | ||
| rs1370627 | chr2 | 101009384 | DUSP2 | 1844 | 0.18 | trans | ||
| rs2014805 | chr2 | 175165192 | DUSP2 | 1844 | 0.01 | trans | ||
| rs9807949 | chr2 | 182655005 | DUSP2 | 1844 | 0.07 | trans | ||
| rs7608942 | chr2 | 182820100 | DUSP2 | 1844 | 4.193E-4 | trans | ||
| rs4674479 | chr2 | 220806451 | DUSP2 | 1844 | 0.01 | trans | ||
| rs1014141 | chr3 | 43810759 | DUSP2 | 1844 | 0.03 | trans | ||
| rs7615639 | chr3 | 43852564 | DUSP2 | 1844 | 0.01 | trans | ||
| rs9809244 | chr3 | 62078922 | DUSP2 | 1844 | 0 | trans | ||
| rs17044826 | chr3 | 66620522 | DUSP2 | 1844 | 0.05 | trans | ||
| rs7637903 | chr3 | 103712766 | DUSP2 | 1844 | 0.14 | trans | ||
| rs6807632 | chr3 | 111395567 | DUSP2 | 1844 | 0.05 | trans | ||
| rs728681 | chr4 | 180794245 | DUSP2 | 1844 | 0.17 | trans | ||
| rs173177 | chr5 | 16161616 | DUSP2 | 1844 | 0.15 | trans | ||
| rs1478417 | chr5 | 122277035 | DUSP2 | 1844 | 0.08 | trans | ||
| rs1977059 | chr6 | 5720825 | DUSP2 | 1844 | 0.15 | trans | ||
| rs6457967 | chr6 | 37002011 | DUSP2 | 1844 | 0.01 | trans | ||
| rs10947885 | chr6 | 40394397 | DUSP2 | 1844 | 0.12 | trans | ||
| rs9450829 | chr6 | 88624436 | DUSP2 | 1844 | 0.12 | trans | ||
| rs6935871 | chr6 | 109107049 | DUSP2 | 1844 | 0.02 | trans | ||
| rs12164028 | chr6 | 136929934 | DUSP2 | 1844 | 0.06 | trans | ||
| rs887719 | chr7 | 21184272 | DUSP2 | 1844 | 0.01 | trans | ||
| rs320082 | chr7 | 29105404 | DUSP2 | 1844 | 6.512E-8 | trans | ||
| rs317720 | chr7 | 29110633 | DUSP2 | 1844 | 5.309E-7 | trans | ||
| rs317714 | chr7 | 29115553 | DUSP2 | 1844 | 2.201E-7 | trans | ||
| rs317724 | chr7 | 29120565 | DUSP2 | 1844 | 5.309E-7 | trans | ||
| rs17141666 | chr7 | 69928490 | DUSP2 | 1844 | 0.13 | trans | ||
| rs12540961 | chr8 | 9689750 | DUSP2 | 1844 | 0.11 | trans | ||
| rs7829733 | chr8 | 9691720 | DUSP2 | 1844 | 5.234E-4 | trans | ||
| rs4242415 | chr8 | 24597175 | DUSP2 | 1844 | 0.15 | trans | ||
| rs16886605 | chr8 | 115894398 | DUSP2 | 1844 | 0.04 | trans | ||
| rs799881 | chr8 | 117260630 | DUSP2 | 1844 | 0.09 | trans | ||
| rs7850851 | chr9 | 37928770 | DUSP2 | 1844 | 0.18 | trans | ||
| rs10121216 | chr9 | 122379089 | DUSP2 | 1844 | 0.18 | trans | ||
| rs10985822 | chr9 | 125685653 | DUSP2 | 1844 | 0.05 | trans | ||
| rs7024546 | chr9 | 125696945 | DUSP2 | 1844 | 0.04 | trans | ||
| rs10985900 | chr9 | 125954348 | DUSP2 | 1844 | 0.11 | trans | ||
| rs2417062 | chr9 | 130715921 | DUSP2 | 1844 | 0.02 | trans | ||
| rs12569493 | chr10 | 74886042 | DUSP2 | 1844 | 0.1 | trans | ||
| rs7092445 | chr10 | 74921991 | DUSP2 | 1844 | 0.15 | trans | ||
| rs11000579 | chr10 | 75027961 | DUSP2 | 1844 | 0.04 | trans | ||
| rs11000607 | chr10 | 75089777 | DUSP2 | 1844 | 0.19 | trans | ||
| rs11203077 | chr10 | 91097084 | DUSP2 | 1844 | 2.026E-5 | trans | ||
| rs10834670 | chr11 | 25404015 | DUSP2 | 1844 | 0.14 | trans | ||
| rs2509929 | chr11 | 58407072 | DUSP2 | 1844 | 0.07 | trans | ||
| rs2186571 | chr11 | 63915699 | DUSP2 | 1844 | 0.16 | trans | ||
| rs1544538 | chr11 | 63988339 | DUSP2 | 1844 | 0.16 | trans | ||
| rs10791832 | chr11 | 65683917 | DUSP2 | 1844 | 0.08 | trans | ||
| rs11052230 | chr12 | 32931283 | DUSP2 | 1844 | 0.02 | trans | ||
| rs11176643 | chr12 | 67629785 | DUSP2 | 1844 | 8.382E-4 | trans | ||
| rs17113079 | chr12 | 74538548 | DUSP2 | 1844 | 0.13 | trans | ||
| rs17113131 | chr12 | 74564573 | DUSP2 | 1844 | 0.13 | trans | ||
| rs17764482 | chr12 | 82483756 | DUSP2 | 1844 | 0.14 | trans | ||
| rs17026331 | chr12 | 97478907 | DUSP2 | 1844 | 0.12 | trans | ||
| rs11066476 | chr12 | 113465366 | DUSP2 | 1844 | 1.066E-4 | trans | ||
| rs4497566 | chr13 | 51694324 | DUSP2 | 1844 | 7.022E-5 | trans | ||
| rs7322001 | chr13 | 94895345 | DUSP2 | 1844 | 0.03 | trans | ||
| snp_a-1952108 | 0 | DUSP2 | 1844 | 0.1 | trans | |||
| rs2993290 | chr13 | 113647189 | DUSP2 | 1844 | 0.15 | trans | ||
| rs17793815 | chr14 | 22794531 | DUSP2 | 1844 | 0.07 | trans | ||
| rs10145685 | chr14 | 22794892 | DUSP2 | 1844 | 0.18 | trans | ||
| rs17793827 | chr14 | 22797560 | DUSP2 | 1844 | 0.09 | trans | ||
| rs10136383 | chr14 | 22801078 | DUSP2 | 1844 | 0.09 | trans | ||
| rs11158850 | chr14 | 70757171 | DUSP2 | 1844 | 0.02 | trans | ||
| rs2444983 | chr15 | 33443919 | DUSP2 | 1844 | 4.942E-4 | trans | ||
| rs2412599 | chr15 | 41347970 | DUSP2 | 1844 | 0.08 | trans | ||
| rs11072201 | chr15 | 70857175 | DUSP2 | 1844 | 0 | trans | ||
| rs17781350 | chr15 | 71108134 | DUSP2 | 1844 | 0.12 | trans | ||
| rs9302779 | chr16 | 3933344 | DUSP2 | 1844 | 0.17 | trans | ||
| rs7200513 | chr16 | 17219474 | DUSP2 | 1844 | 0 | trans | ||
| rs3829495 | chr16 | 17221986 | DUSP2 | 1844 | 5.104E-5 | trans | ||
| rs16949485 | chr16 | 79104374 | DUSP2 | 1844 | 0.13 | trans | ||
| rs2874668 | chr17 | 11631907 | DUSP2 | 1844 | 0.07 | trans | ||
| rs16940752 | chr17 | 11642174 | DUSP2 | 1844 | 0.07 | trans | ||
| rs12452652 | chr17 | 72235560 | DUSP2 | 1844 | 1.586E-9 | trans | ||
| rs17188689 | chr18 | 23359136 | DUSP2 | 1844 | 0.18 | trans | ||
| rs17056775 | chr18 | 72988809 | DUSP2 | 1844 | 0.16 | trans | ||
| rs1523533 | chr20 | 51329866 | DUSP2 | 1844 | 0.01 | trans | ||
| rs2689370 | chr20 | 51335422 | DUSP2 | 1844 | 0.07 | trans | ||
| rs2745070 | chr20 | 51337694 | DUSP2 | 1844 | 0.07 | trans | ||
| rs5992705 | chr22 | 17914003 | DUSP2 | 1844 | 0.03 | trans |
Section II. Transcriptome annotation
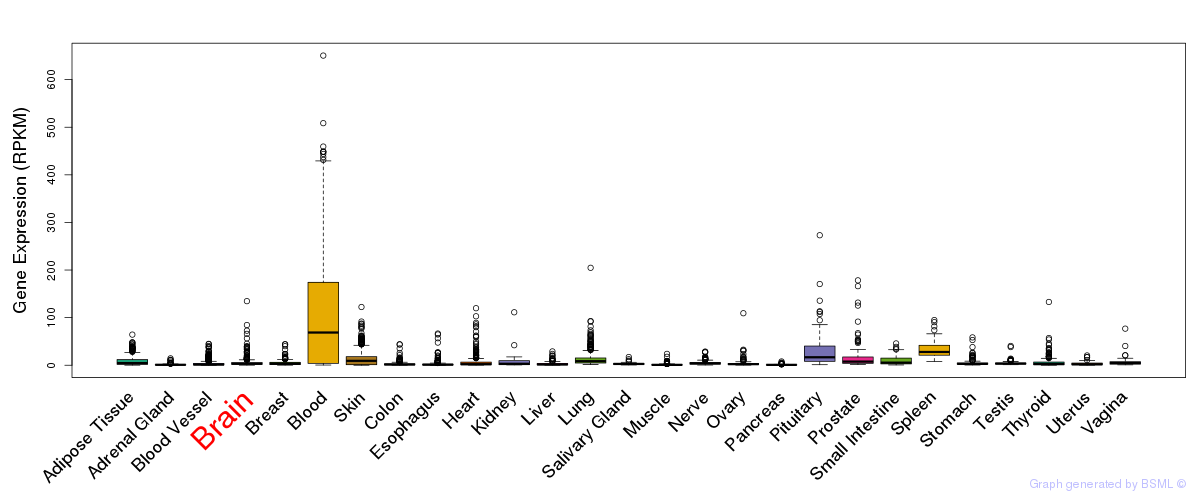
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| UNK | 0.95 | 0.95 |
| LIG1 | 0.95 | 0.90 |
| CPSF1 | 0.94 | 0.94 |
| HAUS5 | 0.94 | 0.93 |
| MCM7 | 0.93 | 0.86 |
| CD276 | 0.93 | 0.91 |
| PLXNB2 | 0.93 | 0.91 |
| RCC1 | 0.93 | 0.83 |
| TCF3 | 0.93 | 0.90 |
| AAAS | 0.93 | 0.91 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| C5orf53 | -0.67 | -0.77 |
| AF347015.31 | -0.66 | -0.85 |
| AF347015.27 | -0.65 | -0.83 |
| MT-CO2 | -0.64 | -0.84 |
| AF347015.33 | -0.62 | -0.79 |
| MT-CYB | -0.62 | -0.81 |
| AIFM3 | -0.61 | -0.66 |
| S100B | -0.61 | -0.73 |
| AF347015.8 | -0.61 | -0.82 |
| IFI27 | -0.60 | -0.75 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004725 | protein tyrosine phosphatase activity | TAS | 7681221 | |
| GO:0016787 | hydrolase activity | IEA | - | |
| GO:0008330 | protein tyrosine/threonine phosphatase activity | TAS | 8107850 | |
| GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity | IEA | - | |
| GO:0051019 | mitogen-activated protein kinase binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000188 | inactivation of MAPK activity | TAS | 8107850 | |
| GO:0006470 | protein amino acid dephosphorylation | IEA | - | |
| GO:0006470 | protein amino acid dephosphorylation | TAS | 8107850 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | TAS | 8107850 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG MAPK SIGNALING PATHWAY | 267 | 205 | All SZGR 2.0 genes in this pathway |
| PICCALUGA ANGIOIMMUNOBLASTIC LYMPHOMA DN | 136 | 86 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE LIVE UP | 485 | 293 | All SZGR 2.0 genes in this pathway |
| OSWALD HEMATOPOIETIC STEM CELL IN COLLAGEN GEL UP | 233 | 161 | All SZGR 2.0 genes in this pathway |
| NAGASHIMA NRG1 SIGNALING UP | 176 | 123 | All SZGR 2.0 genes in this pathway |
| NAGASHIMA EGF SIGNALING UP | 58 | 40 | All SZGR 2.0 genes in this pathway |
| HAHTOLA SEZARY SYNDROM DN | 42 | 26 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE EPIDERMIS DN | 508 | 354 | All SZGR 2.0 genes in this pathway |
| WANG ESOPHAGUS CANCER VS NORMAL UP | 121 | 72 | All SZGR 2.0 genes in this pathway |
| WANG ESOPHAGUS CANCER PROGRESSION UP | 9 | 6 | All SZGR 2.0 genes in this pathway |
| DIRMEIER LMP1 RESPONSE EARLY | 66 | 48 | All SZGR 2.0 genes in this pathway |
| WATTEL AUTONOMOUS THYROID ADENOMA DN | 55 | 29 | All SZGR 2.0 genes in this pathway |
| GROSS HYPOXIA VIA ELK3 DN | 156 | 106 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 48HR DN | 428 | 306 | All SZGR 2.0 genes in this pathway |
| AMIT EGF RESPONSE 40 HELA | 42 | 29 | All SZGR 2.0 genes in this pathway |
| ROSS ACUTE MYELOID LEUKEMIA CBF | 82 | 57 | All SZGR 2.0 genes in this pathway |
| GALINDO IMMUNE RESPONSE TO ENTEROTOXIN | 85 | 67 | All SZGR 2.0 genes in this pathway |
| ZHAN MULTIPLE MYELOMA SPIKED | 22 | 13 | All SZGR 2.0 genes in this pathway |
| BASSO CD40 SIGNALING UP | 101 | 76 | All SZGR 2.0 genes in this pathway |
| WANG CISPLATIN RESPONSE AND XPC DN | 228 | 146 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS 1 | 33 | 24 | All SZGR 2.0 genes in this pathway |
| BILD MYC ONCOGENIC SIGNATURE | 206 | 117 | All SZGR 2.0 genes in this pathway |
| FOSTER TOLERANT MACROPHAGE DN | 409 | 268 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS SILENCED BY METHYLATION UP | 461 | 298 | All SZGR 2.0 genes in this pathway |
| MASSARWEH TAMOXIFEN RESISTANCE DN | 258 | 160 | All SZGR 2.0 genes in this pathway |
| WALLACE PROSTATE CANCER RACE UP | 299 | 167 | All SZGR 2.0 genes in this pathway |
| MARZEC IL2 SIGNALING UP | 115 | 80 | All SZGR 2.0 genes in this pathway |
| RIZKI TUMOR INVASIVENESS 3D UP | 210 | 124 | All SZGR 2.0 genes in this pathway |
| RIZKI TUMOR INVASIVENESS 2D UP | 69 | 46 | All SZGR 2.0 genes in this pathway |
| MITSIADES RESPONSE TO APLIDIN UP | 439 | 257 | All SZGR 2.0 genes in this pathway |
| QI PLASMACYTOMA UP | 259 | 185 | All SZGR 2.0 genes in this pathway |
| VANTVEER BREAST CANCER ESR1 DN | 240 | 153 | All SZGR 2.0 genes in this pathway |
| JISON SICKLE CELL DISEASE DN | 181 | 97 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH INV 16 TRANSLOCATION | 422 | 277 | All SZGR 2.0 genes in this pathway |
| TIAN TNF SIGNALING NOT VIA NFKB | 22 | 16 | All SZGR 2.0 genes in this pathway |
| WONG ADULT TISSUE STEM MODULE | 721 | 492 | All SZGR 2.0 genes in this pathway |
| CHANDRAN METASTASIS DN | 306 | 191 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 30MIN DN | 150 | 99 | All SZGR 2.0 genes in this pathway |
| CHYLA CBFA2T3 TARGETS UP | 387 | 225 | All SZGR 2.0 genes in this pathway |
| BHAT ESR1 TARGETS VIA AKT1 UP | 281 | 183 | All SZGR 2.0 genes in this pathway |
| IVANOVSKA MIR106B TARGETS | 90 | 56 | All SZGR 2.0 genes in this pathway |
| KASLER HDAC7 TARGETS 1 UP | 194 | 133 | All SZGR 2.0 genes in this pathway |
| TORCHIA TARGETS OF EWSR1 FLI1 FUSION DN | 321 | 200 | All SZGR 2.0 genes in this pathway |
| WANG MLL TARGETS | 289 | 188 | All SZGR 2.0 genes in this pathway |
| ALFANO MYC TARGETS | 239 | 156 | All SZGR 2.0 genes in this pathway |
| FORTSCHEGGER PHF8 TARGETS UP | 279 | 155 | All SZGR 2.0 genes in this pathway |
| VANOEVELEN MYOGENESIS SIN3A TARGETS | 220 | 133 | All SZGR 2.0 genes in this pathway |
| HOLLEMAN DAUNORUBICIN ALL UP | 7 | 5 | All SZGR 2.0 genes in this pathway |
| SMIRNOV RESPONSE TO IR 6HR DN | 114 | 69 | All SZGR 2.0 genes in this pathway |
| GHANDHI DIRECT IRRADIATION UP | 110 | 68 | All SZGR 2.0 genes in this pathway |
| GHANDHI BYSTANDER IRRADIATION UP | 86 | 54 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 2ND EGF PULSE ONLY | 1725 | 838 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-142-5p | 592 | 599 | 1A,m8 | hsa-miR-142-5p | CAUAAAGUAGAAAGCACUAC |
| miR-17-5p/20/93.mr/106/519.d | 590 | 597 | 1A,m8 | hsa-miR-17-5p | CAAAGUGCUUACAGUGCAGGUAGU |
| hsa-miR-20abrain | UAAAGUGCUUAUAGUGCAGGUAG | ||||
| hsa-miR-106a | AAAAGUGCUUACAGUGCAGGUAGC | ||||
| hsa-miR-106bSZ | UAAAGUGCUGACAGUGCAGAU | ||||
| hsa-miR-20bSZ | CAAAGUGCUCAUAGUGCAGGUAG | ||||
| hsa-miR-519d | CAAAGUGCCUCCCUUUAGAGUGU | ||||
| miR-29 | 178 | 184 | m8 | hsa-miR-29aSZ | UAGCACCAUCUGAAAUCGGUU |
| hsa-miR-29bSZ | UAGCACCAUUUGAAAUCAGUGUU | ||||
| hsa-miR-29cSZ | UAGCACCAUUUGAAAUCGGU | ||||
| miR-93.hd/291-3p/294/295/302/372/373/520 | 589 | 595 | m8 | hsa-miR-93brain | AAAGUGCUGUUCGUGCAGGUAG |
| hsa-miR-302a | UAAGUGCUUCCAUGUUUUGGUGA | ||||
| hsa-miR-302b | UAAGUGCUUCCAUGUUUUAGUAG | ||||
| hsa-miR-302c | UAAGUGCUUCCAUGUUUCAGUGG | ||||
| hsa-miR-302d | UAAGUGCUUCCAUGUUUGAGUGU | ||||
| hsa-miR-372 | AAAGUGCUGCGACAUUUGAGCGU | ||||
| hsa-miR-373 | GAAGUGCUUCGAUUUUGGGGUGU | ||||
| hsa-miR-520e | AAAGUGCUUCCUUUUUGAGGG | ||||
| hsa-miR-520a | AAAGUGCUUCCCUUUGGACUGU | ||||
| hsa-miR-520b | AAAGUGCUUCCUUUUAGAGGG | ||||
| hsa-miR-520c | AAAGUGCUUCCUUUUAGAGGGUU | ||||
| hsa-miR-520d | AAAGUGCUUCUCUUUGGUGGGUU | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.