Gene Page: A2M
Summary ?
| GeneID | 2 |
| Symbol | A2M |
| Synonyms | A2MD|CPAMD5|FWP007|S863-7 |
| Description | alpha-2-macroglobulin |
| Reference | MIM:103950|HGNC:HGNC:7|Ensembl:ENSG00000175899|HPRD:00072|Vega:OTTHUMG00000150267 |
| Gene type | protein-coding |
| Map location | 12p13.31 |
| Pascal p-value | 0.529 |
| Sherlock p-value | 0.668 |
| Fetal beta | -1.656 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 4.179 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg27166707 | 12 | 9227209 | A2M | 7.16E-5 | 0.404 | 0.025 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs1999514 | chr13 | 84147878 | A2M | 2 | 0.08 | trans |
Section II. Transcriptome annotation
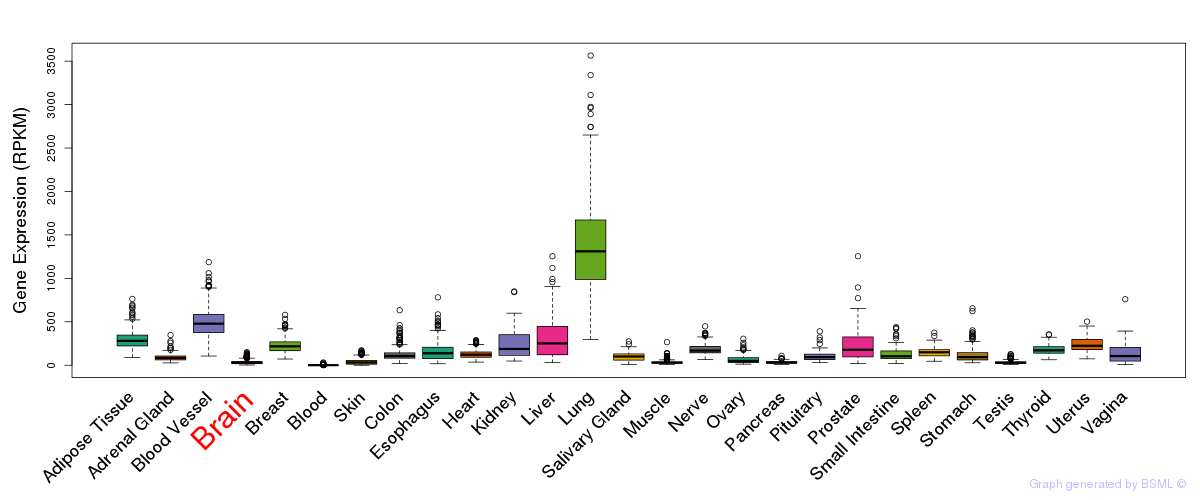
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| NDUFB7 | 0.88 | 0.47 |
| LSMD1 | 0.87 | 0.51 |
| C17orf90 | 0.85 | 0.61 |
| ZNF593 | 0.84 | 0.58 |
| C7orf59 | 0.83 | 0.58 |
| C19orf60 | 0.83 | 0.70 |
| HBQ1 | 0.83 | 0.62 |
| C9orf16 | 0.83 | 0.55 |
| PSMB10 | 0.82 | 0.50 |
| NDUFA2 | 0.82 | 0.49 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| CANX | -0.39 | -0.37 |
| AC005332.1 | -0.38 | -0.30 |
| ARHGAP5 | -0.38 | -0.40 |
| DYNC1LI2 | -0.37 | -0.30 |
| ZNHIT6 | -0.37 | -0.34 |
| KIAA1033 | -0.37 | -0.37 |
| RAB5A | -0.37 | -0.32 |
| XPOT | -0.36 | -0.29 |
| CD2AP | -0.36 | -0.36 |
| YES1 | -0.36 | -0.43 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004867 | serine-type endopeptidase inhibitor activity | IEA | - | |
| GO:0017114 | wide-spectrum protease inhibitor activity | IEA | - | |
| GO:0019959 | interleukin-8 binding | IPI | 10880251 | |
| GO:0019966 | interleukin-1 binding | IDA | 9714181 | |
| GO:0019899 | enzyme binding | IPI | 11435418 | |
| GO:0043120 | tumor necrosis factor binding | IDA | 9714181 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0051260 | protein homooligomerization | NAS | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005576 | extracellular region | EXP | 59727 | |
| GO:0005576 | extracellular region | NAS | 14718574 | |
| GO:0005634 | nucleus | IDA | 18029348 | |
| GO:0005737 | cytoplasm | IDA | 18029348 | |
| GO:0031093 | platelet alpha granule lumen | EXP | 59727 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ADAM19 | FKSG34 | MADDAM | MLTNB | ADAM metallopeptidase domain 19 (meltrin beta) | - | HPRD | 11162584 |
| ADAMTS1 | C3-C5 | KIAA1346 | METH1 | ADAM metallopeptidase with thrombospondin type 1 motif, 1 | - | HPRD,BioGRID | 10373500 |
| AMBP | EDC1 | HCP | HI30 | IATIL | ITI | ITIL | ITILC | UTI | alpha-1-microglobulin/bikunin precursor | - | HPRD,BioGRID | 1697852 |7519849 |7533162 |
| APOE | AD2 | LPG | MGC1571 | apoprotein | apolipoprotein E | - | HPRD,BioGRID | 9831625 |
| B2M | - | beta-2-microglobulin | - | HPRD,BioGRID | 10731476 |
| CPB2 | CPU | PCPB | TAFI | carboxypeptidase B2 (plasma) | Reconstituted Complex | BioGRID | 8662763 |
| CTSB | APPS | CPSB | cathepsin B | - | HPRD | 2476070 |
| ELA1 | - | elastase 1, pancreatic | - | HPRD | 6153632 |6191979 |
| HSPA5 | BIP | FLJ26106 | GRP78 | MIF2 | heat shock 70kDa protein 5 (glucose-regulated protein, 78kDa) | - | HPRD,BioGRID | 12194978 |
| IL10 | CSIF | IL-10 | IL10A | MGC126450 | MGC126451 | TGIF | interleukin 10 | - | HPRD,BioGRID | 10714547 |
| IL1B | IL-1 | IL1-BETA | IL1F2 | interleukin 1, beta | - | HPRD | 9714181 |
| IL4 | BCGF-1 | BCGF1 | BSF1 | IL-4 | MGC79402 | interleukin 4 | Reconstituted Complex | BioGRID | 10714547 |
| KLK13 | DKFZp586J1923 | KLK-L4 | KLKL4 | kallikrein-related peptidase 13 | Reconstituted Complex | BioGRID | 14687906 |
| KLK3 | APS | KLK2A1 | PSA | hK3 | kallikrein-related peptidase 3 | - | HPRD | 8583572 |
| KLK3 | APS | KLK2A1 | PSA | hK3 | kallikrein-related peptidase 3 | Affinity Capture-Western | BioGRID | 1702714 |
| LCAT | - | lecithin-cholesterol acyltransferase | - | HPRD,BioGRID | 11435418 |
| LEP | FLJ94114 | OB | OBS | leptin | - | HPRD,BioGRID | 9724081 |
| LRP1 | A2MR | APOER | APR | CD91 | FLJ16451 | IGFBP3R | LRP | MGC88725 | TGFBR5 | low density lipoprotein-related protein 1 (alpha-2-macroglobulin receptor) | - | HPRD,BioGRID | 10652313 |
| MMP2 | CLG4 | CLG4A | MMP-II | MONA | TBE-1 | matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase) | Reconstituted Complex | BioGRID | 9344465 |
| NGF | Beta-NGF | HSAN5 | MGC161426 | MGC161428 | NGFB | nerve growth factor (beta polypeptide) | - | HPRD,BioGRID | 10681572 |
| PAEP | GD | GdA | GdF | GdS | MGC138509 | MGC142288 | PAEG | PEP | PP14 | progestagen-associated endometrial protein | - | HPRD,BioGRID | 11023837 |
| PDGFB | FLJ12858 | PDGF2 | SIS | SSV | c-sis | platelet-derived growth factor beta polypeptide (simian sarcoma viral (v-sis) oncogene homolog) | - | HPRD | 10681572 |
| STAT3 | APRF | FLJ20882 | HIES | MGC16063 | signal transducer and activator of transcription 3 (acute-phase response factor) | An unspecified isoform of STAT3 binds APRE. | BIND | 15950906 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG COMPLEMENT AND COAGULATION CASCADES | 69 | 49 | All SZGR 2.0 genes in this pathway |
| PID IL6 7 PATHWAY | 47 | 40 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY RHO GTPASES | 113 | 81 | All SZGR 2.0 genes in this pathway |
| REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | 89 | 56 | All SZGR 2.0 genes in this pathway |
| REACTOME HDL MEDIATED LIPID TRANSPORT | 15 | 12 | All SZGR 2.0 genes in this pathway |
| REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | 32 | 22 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF LIPIDS AND LIPOPROTEINS | 478 | 302 | All SZGR 2.0 genes in this pathway |
| REACTOME INTRINSIC PATHWAY | 17 | 12 | All SZGR 2.0 genes in this pathway |
| REACTOME LIPID DIGESTION MOBILIZATION AND TRANSPORT | 46 | 37 | All SZGR 2.0 genes in this pathway |
| REACTOME HEMOSTASIS | 466 | 331 | All SZGR 2.0 genes in this pathway |
| REACTOME LIPOPROTEIN METABOLISM | 28 | 24 | All SZGR 2.0 genes in this pathway |
| REACTOME PLATELET ACTIVATION SIGNALING AND AGGREGATION | 208 | 138 | All SZGR 2.0 genes in this pathway |
| PICCALUGA ANGIOIMMUNOBLASTIC LYMPHOMA UP | 205 | 140 | All SZGR 2.0 genes in this pathway |
| KORKOLA TERATOMA UP | 16 | 11 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 10D DN | 142 | 90 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION DN | 329 | 219 | All SZGR 2.0 genes in this pathway |
| TANG SENESCENCE TP53 TARGETS UP | 33 | 20 | All SZGR 2.0 genes in this pathway |
| HU ANGIOGENESIS DN | 37 | 25 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 2B | 392 | 251 | All SZGR 2.0 genes in this pathway |
| HERNANDEZ ABERRANT MITOSIS BY DOCETACEL 2NM DN | 25 | 18 | All SZGR 2.0 genes in this pathway |
| HERNANDEZ MITOTIC ARREST BY DOCETAXEL 2 DN | 19 | 13 | All SZGR 2.0 genes in this pathway |
| MARTORIATI MDM4 TARGETS FETAL LIVER UP | 227 | 137 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT AND CANCER BOX1 UP | 15 | 10 | All SZGR 2.0 genes in this pathway |
| COLIN PILOCYTIC ASTROCYTOMA VS GLIOBLASTOMA DN | 28 | 21 | All SZGR 2.0 genes in this pathway |
| BENPORATH CYCLING GENES | 648 | 385 | All SZGR 2.0 genes in this pathway |
| BENPORATH PROLIFERATION | 147 | 80 | All SZGR 2.0 genes in this pathway |
| CERVERA SDHB TARGETS 2 | 114 | 76 | All SZGR 2.0 genes in this pathway |
| CHESLER BRAIN D6MIT150 QTL CIS | 7 | 6 | All SZGR 2.0 genes in this pathway |
| SMITH TERT TARGETS DN | 87 | 69 | All SZGR 2.0 genes in this pathway |
| NING CHRONIC OBSTRUCTIVE PULMONARY DISEASE UP | 157 | 105 | All SZGR 2.0 genes in this pathway |
| COLLER MYC TARGETS DN | 9 | 5 | All SZGR 2.0 genes in this pathway |
| LENAOUR DENDRITIC CELL MATURATION UP | 114 | 84 | All SZGR 2.0 genes in this pathway |
| TAVOR CEBPA TARGETS DN | 30 | 21 | All SZGR 2.0 genes in this pathway |
| ABRAHAM ALPC VS MULTIPLE MYELOMA UP | 26 | 22 | All SZGR 2.0 genes in this pathway |
| KANG IMMORTALIZED BY TERT DN | 102 | 67 | All SZGR 2.0 genes in this pathway |
| ZHAN MULTIPLE MYELOMA DN | 41 | 27 | All SZGR 2.0 genes in this pathway |
| JOSEPH RESPONSE TO SODIUM BUTYRATE DN | 64 | 45 | All SZGR 2.0 genes in this pathway |
| LINDVALL IMMORTALIZED BY TERT UP | 78 | 48 | All SZGR 2.0 genes in this pathway |
| HENDRICKS SMARCA4 TARGETS DN | 53 | 34 | All SZGR 2.0 genes in this pathway |
| RODWELL AGING KIDNEY NO BLOOD UP | 222 | 139 | All SZGR 2.0 genes in this pathway |
| RODWELL AGING KIDNEY UP | 487 | 303 | All SZGR 2.0 genes in this pathway |
| ABE VEGFA TARGETS | 20 | 15 | All SZGR 2.0 genes in this pathway |
| JIANG HYPOXIA NORMAL | 311 | 205 | All SZGR 2.0 genes in this pathway |
| WANG CISPLATIN RESPONSE AND XPC UP | 202 | 115 | All SZGR 2.0 genes in this pathway |
| ZHONG SECRETOME OF LUNG CANCER AND ENDOTHELIUM | 66 | 47 | All SZGR 2.0 genes in this pathway |
| RIGGI EWING SARCOMA PROGENITOR UP | 430 | 288 | All SZGR 2.0 genes in this pathway |
| ACEVEDO NORMAL TISSUE ADJACENT TO LIVER TUMOR UP | 174 | 96 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER TUMOR VS NORMAL ADJACENT TISSUE DN | 274 | 165 | All SZGR 2.0 genes in this pathway |
| ZHANG BREAST CANCER PROGENITORS DN | 145 | 93 | All SZGR 2.0 genes in this pathway |
| ZHENG GLIOBLASTOMA PLASTICITY UP | 250 | 168 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL CULTURED VS FRESH UP | 425 | 298 | All SZGR 2.0 genes in this pathway |
| RAY TUMORIGENESIS BY ERBB2 CDC25A UP | 104 | 57 | All SZGR 2.0 genes in this pathway |
| YAUCH HEDGEHOG SIGNALING PARACRINE DN | 264 | 159 | All SZGR 2.0 genes in this pathway |
| TSAI RESPONSE TO RADIATION THERAPY | 32 | 20 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| RUIZ TNC TARGETS UP | 153 | 107 | All SZGR 2.0 genes in this pathway |
| CAIRO LIVER DEVELOPMENT DN | 222 | 141 | All SZGR 2.0 genes in this pathway |
| DANG REGULATED BY MYC DN | 253 | 192 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN ES ICP WITH H3K4ME3 | 718 | 401 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN NPC ICP WITH H3K4ME3 | 445 | 257 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS UP | 504 | 321 | All SZGR 2.0 genes in this pathway |
| ISSAEVA MLL2 TARGETS | 62 | 35 | All SZGR 2.0 genes in this pathway |
| YANG BCL3 TARGETS UP | 364 | 236 | All SZGR 2.0 genes in this pathway |
| FORTSCHEGGER PHF8 TARGETS UP | 279 | 155 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 2ND EGF PULSE ONLY | 1725 | 838 | All SZGR 2.0 genes in this pathway |
| NABA ECM REGULATORS | 238 | 125 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME ASSOCIATED | 753 | 411 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME | 1028 | 559 | All SZGR 2.0 genes in this pathway |