Gene Page: EPB41
Summary ?
| GeneID | 2035 |
| Symbol | EPB41 |
| Synonyms | 4.1R|EL1|HE |
| Description | erythrocyte membrane protein band 4.1 |
| Reference | MIM:130500|HGNC:HGNC:3377|Ensembl:ENSG00000159023|HPRD:00558|Vega:OTTHUMG00000003644 |
| Gene type | protein-coding |
| Map location | 1p33-p32 |
| Pascal p-value | 8.17E-5 |
| Fetal beta | 2.665 |
| DMG | 1 (# studies) |
| eGene | Cerebellum |
| Support | CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| DNM:Fromer_2014 | Whole Exome Sequencing analysis | This study reported a WES study of 623 schizophrenia trios, reporting DNMs using genomic DNA. | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 3.9116 |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| EPB41 | chr1 | 29344918 | T | TTGA | NM_001166005 NM_001166006 NM_001166007 NM_004437 NM_203342 NM_203343 | . . . . . . | out-of-frame-codon-insertion out-of-frame-codon-insertion out-of-frame-codon-insertion out-of-frame-codon-insertion out-of-frame-codon-insertion out-of-frame-codon-insertion | Schizophrenia | DNM:Fromer_2014 |
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg07154196 | 1 | 29214064 | EPB41 | 1.895E-4 | -0.338 | 0.034 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
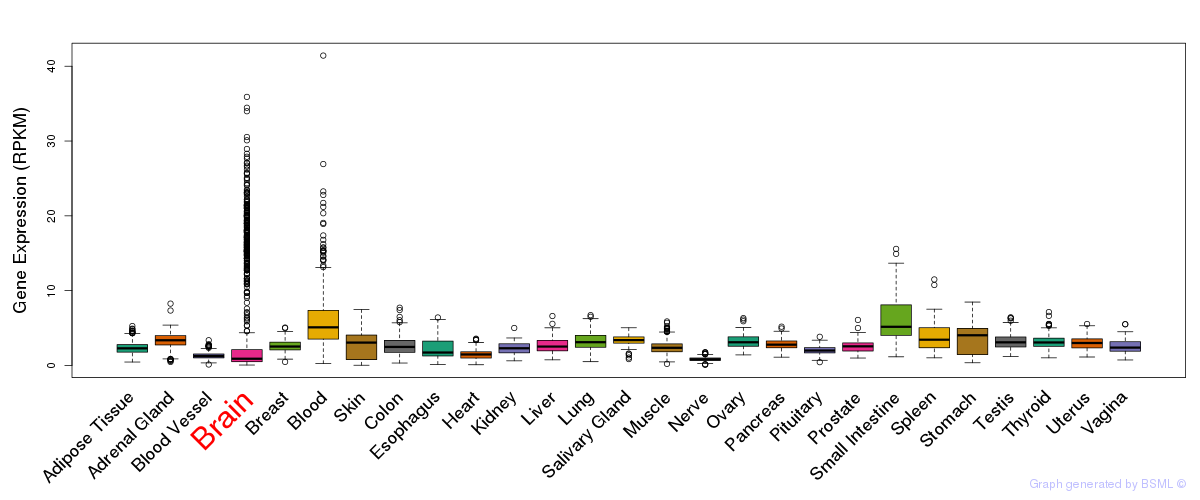
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| PTBP1 | 0.93 | 0.82 |
| TGIF2 | 0.92 | 0.73 |
| SMO | 0.90 | 0.80 |
| TP53 | 0.89 | 0.73 |
| ITPRIP | 0.89 | 0.74 |
| CHST14 | 0.89 | 0.79 |
| ERBB2 | 0.88 | 0.81 |
| TEAD2 | 0.88 | 0.66 |
| TCF19 | 0.88 | 0.61 |
| CDK2 | 0.88 | 0.66 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| C5orf53 | -0.46 | -0.53 |
| CCNI2 | -0.41 | -0.45 |
| CHN1 | -0.41 | -0.48 |
| OMG | -0.41 | -0.48 |
| CA11 | -0.40 | -0.46 |
| SERPINI1 | -0.40 | -0.49 |
| EPHX4 | -0.40 | -0.51 |
| CKMT1A | -0.40 | -0.45 |
| ACOT13 | -0.40 | -0.50 |
| ACYP2 | -0.39 | -0.45 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003779 | actin binding | IEA | - | |
| GO:0005545 | phosphatidylinositol binding | IDA | 16669616 | |
| GO:0005488 | binding | IEA | - | |
| GO:0005198 | structural molecule activity | IEA | - | |
| GO:0005200 | structural constituent of cytoskeleton | TAS | 6894932 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0008015 | blood circulation | TAS | 6894932 | |
| GO:0030866 | cortical actin cytoskeleton organization | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005856 | cytoskeleton | IEA | - | |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0008091 | spectrin | TAS | 6894932 | |
| GO:0005886 | plasma membrane | TAS | 9598318 | |
| GO:0019898 | extrinsic to membrane | IEA | - | |
| GO:0043234 | protein complex | IDA | 16060676 | |
| GO:0030863 | cortical cytoskeleton | IDA | 16254212 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ACTA1 | ACTA | ASMA | CFTD | CFTD1 | CFTDM | MPFD | NEM1 | NEM2 | NEM3 | actin, alpha 1, skeletal muscle | - | HPRD | 11071908 |
| CALR | CRT | FLJ26680 | RO | SSA | cC1qR | calreticulin | Affinity Capture-MS | BioGRID | 17353931 |
| CASK | CAGH39 | CMG | FLJ22219 | FLJ31914 | LIN2 | MICPCH | TNRC8 | calcium/calmodulin-dependent serine protein kinase (MAGUK family) | - | HPRD,BioGRID | 9660868 |
| CD44 | CDW44 | CSPG8 | ECMR-III | HCELL | IN | LHR | MC56 | MDU2 | MDU3 | MGC10468 | MIC4 | MUTCH-I | Pgp1 | CD44 molecule (Indian blood group) | - | HPRD | 12901833 |
| CDK2 | p33(CDK2) | cyclin-dependent kinase 2 | Affinity Capture-MS | BioGRID | 17353931 |
| CENPJ | BM032 | CPAP | LAP | LIP1 | MCPH6 | MGC131581 | MGC131582 | MGC142222 | MGC142224 | centromere protein J | - | HPRD,BioGRID | 11003675 |
| DCTN1 | DAP-150 | DP-150 | HMN7B | P135 | dynactin 1 (p150, glued homolog, Drosophila) | Affinity Capture-Western | BioGRID | 10189366 |
| DLG1 | DKFZp761P0818 | DKFZp781B0426 | DLGH1 | SAP97 | dJ1061C18.1.1 | hdlg | discs, large homolog 1 (Drosophila) | - | HPRD,BioGRID | 7937897 |
| DRD2 | D2DR | D2R | dopamine receptor D2 | - | HPRD | 12181426 |
| DRD3 | D3DR | ETM1 | FET1 | MGC149204 | MGC149205 | dopamine receptor D3 | - | HPRD | 12181426 |
| DYNC1H1 | DHC1 | DHC1a | DKFZp686P2245 | DNCH1 | DNCL | DNECL | DYHC | Dnchc1 | HL-3 | KIAA0325 | p22 | dynein, cytoplasmic 1, heavy chain 1 | Affinity Capture-Western | BioGRID | 10189366 |
| ECHS1 | SCEH | enoyl Coenzyme A hydratase, short chain, 1, mitochondrial | Affinity Capture-MS | BioGRID | 17353931 |
| EIF3G | EIF3-P42 | EIF3S4 | eIF3-delta | eIF3-p44 | eukaryotic translation initiation factor 3, subunit G | - | HPRD,BioGRID | 10887144 |
| EPB42 | MGC116735 | MGC116737 | PA | erythrocyte membrane protein band 4.2 | - | HPRD | 2968981 |
| GYPC | CD236 | CD236R | GE | GPC | GYPD | MGC117309 | MGC126191 | MGC126192 | glycophorin C (Gerbich blood group) | - | HPRD | 11423550 |
| KPNA2 | IPOA1 | QIP2 | RCH1 | SRP1alpha | karyopherin alpha 2 (RAG cohort 1, importin alpha 1) | - | HPRD,BioGRID | 10359596 |
| MYH9 | DFNA17 | EPSTS | FTNS | MGC104539 | MHA | NMHC-II-A | NMMHCA | myosin, heavy chain 9, non-muscle | Affinity Capture-MS | BioGRID | 17353931 |
| MYL1 | MLC1F | MLC3F | myosin, light chain 1, alkali; skeletal, fast | - | HPRD | 11071908 |
| NPPA | ANF | ANP | ATFB6 | CDD-ANF | PND | natriuretic peptide precursor A | Reconstituted Complex | BioGRID | 12542678 |
| NUMA1 | NUMA | nuclear mitotic apparatus protein 1 | - | HPRD,BioGRID | 10189366 |
| PAICS | ADE2 | ADE2H1 | AIRC | DKFZp781N1372 | MGC1343 | MGC5024 | PAIS | phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase | Affinity Capture-MS | BioGRID | 17353931 |
| PFAS | FGAMS | FGARAT | KIAA0361 | PURL | phosphoribosylformylglycinamidine synthase | Affinity Capture-MS | BioGRID | 17353931 |
| SLC4A1 | AE1 | BND3 | CD233 | DI | EMPB3 | EPB3 | FR | MGC116750 | MGC116753 | MGC126619 | MGC126623 | RTA1A | SW | WD | WD1 | WR | solute carrier family 4, anion exchanger, member 1 (erythrocyte membrane protein band 3, Diego blood group) | - | HPRD | 1639060 |
| SPTAN1 | (ALPHA)II-SPECTRIN | FLJ44613 | NEAS | spectrin, alpha, non-erythrocytic 1 (alpha-fodrin) | Reconstituted Complex | BioGRID | 12044158 |
| SPTAN1 | (ALPHA)II-SPECTRIN | FLJ44613 | NEAS | spectrin, alpha, non-erythrocytic 1 (alpha-fodrin) | - | HPRD | 12049649 |
| SPTB | HSpTB1 | spectrin, beta, erythrocytic | - | HPRD,BioGRID | 7673158 |
| SPTBN1 | ELF | SPTB2 | betaSpII | spectrin, beta, non-erythrocytic 1 | - | HPRD | 12901833 |
| TAGLN2 | HA1756 | KIAA0120 | transgelin 2 | Affinity Capture-MS | BioGRID | 17353931 |
| TJP2 | MGC26306 | X104 | ZO-2 | ZO2 | tight junction protein 2 (zona occludens 2) | - | HPRD,BioGRID | 10874042 |
| TPM1 | C15orf13 | CMD1Y | HTM-alpha | TMSA | tropomyosin 1 (alpha) | - | HPRD | 11071908 |
| U2AF1 | DKFZp313J1712 | FP793 | RN | RNU2AF1 | U2AF35 | U2AFBP | U2 small nuclear RNA auxiliary factor 1 | - | HPRD | 9645944 |
| VARS | G7A | VARS1 | VARS2 | valyl-tRNA synthetase | Affinity Capture-MS | BioGRID | 17353931 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG TIGHT JUNCTION | 134 | 86 | All SZGR 2.0 genes in this pathway |
| PID SYNDECAN 2 PATHWAY | 33 | 27 | All SZGR 2.0 genes in this pathway |
| BERTUCCI MEDULLARY VS DUCTAL BREAST CANCER UP | 206 | 111 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE LIVE UP | 485 | 293 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| DEURIG T CELL PROLYMPHOCYTIC LEUKEMIA UP | 368 | 234 | All SZGR 2.0 genes in this pathway |
| BOGNI TREATMENT RELATED MYELOID LEUKEMIA UP | 30 | 19 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE LPS UP | 431 | 237 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION UP | 1278 | 748 | All SZGR 2.0 genes in this pathway |
| CONCANNON APOPTOSIS BY EPOXOMICIN UP | 239 | 157 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS G UP | 238 | 135 | All SZGR 2.0 genes in this pathway |
| LASTOWSKA NEUROBLASTOMA COPY NUMBER DN | 800 | 473 | All SZGR 2.0 genes in this pathway |
| PEREZ TP53 TARGETS | 1174 | 695 | All SZGR 2.0 genes in this pathway |
| RASHI RESPONSE TO IONIZING RADIATION 5 | 147 | 89 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| PUJANA CHEK2 PCC NETWORK | 779 | 480 | All SZGR 2.0 genes in this pathway |
| WEI MIR34A TARGETS | 148 | 97 | All SZGR 2.0 genes in this pathway |
| RICKMAN TUMOR DIFFERENTIATED WELL VS POORLY UP | 236 | 139 | All SZGR 2.0 genes in this pathway |
| SAKAI TUMOR INFILTRATING MONOCYTES DN | 81 | 51 | All SZGR 2.0 genes in this pathway |
| SU PANCREAS | 54 | 30 | All SZGR 2.0 genes in this pathway |
| MCCLUNG CREB1 TARGETS DN | 57 | 39 | All SZGR 2.0 genes in this pathway |
| ULE SPLICING VIA NOVA2 | 43 | 38 | All SZGR 2.0 genes in this pathway |
| MCCLUNG COCAIN REWARD 4WK | 75 | 47 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 6HR DN | 160 | 101 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RESPONSE TO TRABECTEDIN DN | 271 | 175 | All SZGR 2.0 genes in this pathway |
| ZHENG BOUND BY FOXP3 | 491 | 310 | All SZGR 2.0 genes in this pathway |
| ZHENG FOXP3 TARGETS IN THYMUS DN | 12 | 9 | All SZGR 2.0 genes in this pathway |
| SANSOM APC MYC TARGETS | 217 | 138 | All SZGR 2.0 genes in this pathway |
| RIZKI TUMOR INVASIVENESS 3D DN | 270 | 181 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS DN | 435 | 289 | All SZGR 2.0 genes in this pathway |
| LEE DIFFERENTIATING T LYMPHOCYTE | 200 | 115 | All SZGR 2.0 genes in this pathway |
| HAN SATB1 TARGETS UP | 395 | 249 | All SZGR 2.0 genes in this pathway |
| CHEN METABOLIC SYNDROM NETWORK | 1210 | 725 | All SZGR 2.0 genes in this pathway |
| VALK AML CLUSTER 7 | 28 | 16 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH T 8 21 TRANSLOCATION | 368 | 247 | All SZGR 2.0 genes in this pathway |
| CHANDRAN METASTASIS UP | 221 | 135 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 11 | 103 | 68 | All SZGR 2.0 genes in this pathway |
| VERHAAK GLIOBLASTOMA PRONEURAL | 177 | 132 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 24HR DN | 505 | 328 | All SZGR 2.0 genes in this pathway |
| WIERENGA STAT5A TARGETS UP | 217 | 131 | All SZGR 2.0 genes in this pathway |
| MIYAGAWA TARGETS OF EWSR1 ETS FUSIONS UP | 259 | 159 | All SZGR 2.0 genes in this pathway |
| STEINER ERYTHROCYTE MEMBRANE GENES | 15 | 10 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY STEM CELL DN | 428 | 246 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-128 | 703 | 709 | 1A | hsa-miR-128a | UCACAGUGAACCGGUCUCUUUU |
| hsa-miR-128b | UCACAGUGAACCGGUCUCUUUC | ||||
| miR-144 | 701 | 707 | m8 | hsa-miR-144 | UACAGUAUAGAUGAUGUACUAG |
| miR-205 | 2991 | 2998 | 1A,m8 | hsa-miR-205 | UCCUUCAUUCCACCGGAGUCUG |
| miR-27 | 703 | 710 | 1A,m8 | hsa-miR-27abrain | UUCACAGUGGCUAAGUUCCGC |
| hsa-miR-27bbrain | UUCACAGUGGCUAAGUUCUGC | ||||
| miR-30-5p | 3042 | 3049 | 1A,m8 | hsa-miR-30a-5p | UGUAAACAUCCUCGACUGGAAG |
| hsa-miR-30cbrain | UGUAAACAUCCUACACUCUCAGC | ||||
| hsa-miR-30dSZ | UGUAAACAUCCCCGACUGGAAG | ||||
| hsa-miR-30bSZ | UGUAAACAUCCUACACUCAGCU | ||||
| hsa-miR-30e-5p | UGUAAACAUCCUUGACUGGA | ||||
| miR-331 | 944 | 950 | 1A | hsa-miR-331brain | GCCCCUGGGCCUAUCCUAGAA |
| miR-377 | 2495 | 2502 | 1A,m8 | hsa-miR-377 | AUCACACAAAGGCAACUUUUGU |
| miR-381 | 3190 | 3196 | m8 | hsa-miR-381 | UAUACAAGGGCAAGCUCUCUGU |
| miR-504 | 2560 | 2567 | 1A,m8 | hsa-miR-504 | AGACCCUGGUCUGCACUCUAU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.