Gene Page: F10
Summary ?
| GeneID | 2159 |
| Symbol | F10 |
| Synonyms | FX|FXA |
| Description | coagulation factor X |
| Reference | MIM:613872|HGNC:HGNC:3528|Ensembl:ENSG00000126218|HPRD:01966|Vega:OTTHUMG00000017374 |
| Gene type | protein-coding |
| Map location | 13q34 |
| Pascal p-value | 0.168 |
| Fetal beta | -0.939 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenics,schizophrenias,schizotypal | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0033 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
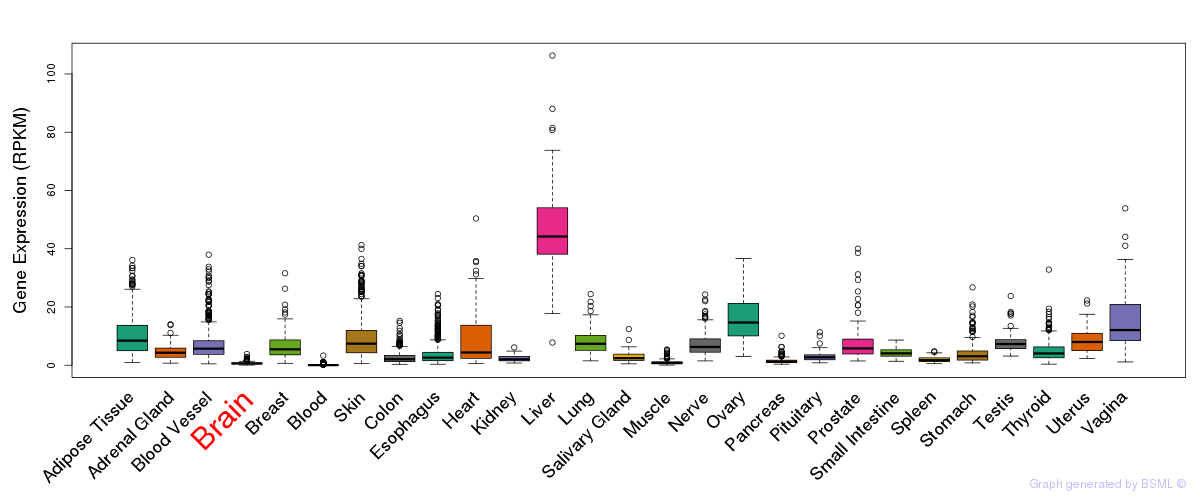
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004252 | serine-type endopeptidase activity | TAS | glutamate (GO term level: 7) | 3011603 |
| GO:0005509 | calcium ion binding | IEA | - | |
| GO:0005515 | protein binding | IPI | 18267072 | |
| GO:0008233 | peptidase activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006508 | proteolysis | IEA | - | |
| GO:0007598 | blood coagulation, extrinsic pathway | EXP | 7598447 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005576 | extracellular region | EXP | 2110473 |2271516 |3052293 |8639673 | |
| GO:0005576 | extracellular region | NAS | 14718574 | |
| GO:0005886 | plasma membrane | EXP | 2271516 |3052293 |14982929 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| F10 | FX | FXA | coagulation factor X | - | HPRD | 8355279 |8663221 |8939944 |
| F3 | CD142 | TF | TFA | coagulation factor III (thromboplastin, tissue factor) | - | HPRD | 8702971 |
| F5 | FVL | PCCF | coagulation factor V (proaccelerin, labile factor) | - | HPRD,BioGRID | 2303476 |
| F7 | - | coagulation factor VII (serum prothrombin conversion accelerator) | - | HPRD,BioGRID | 11560488 |
| F8 | AHF | DXS1253E | F8B | F8C | FVIII | HEMA | coagulation factor VIII, procoagulant component | - | HPRD,BioGRID | 9759621 |10521497 |
| PLAT | DKFZp686I03148 | T-PA | TPA | plasminogen activator, tissue | - | HPRD,BioGRID | 11371191 |
| PLG | DKFZp779M0222 | plasminogen | - | HPRD,BioGRID | 11371191 |
| PRKAB1 | AMPK | HAMPKb | MGC17785 | protein kinase, AMP-activated, beta 1 non-catalytic subunit | Two-hybrid | BioGRID | 16169070 |
| PROS1 | PROS | PS21 | PS22 | PS23 | PS24 | PS25 | PSA | protein S (alpha) | - | HPRD | 8146182 |
| SERPINA5 | PAI3 | PCI | PLANH3 | PROCI | serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 5 | - | HPRD | 2551064 |
| SERPINB6 | CAP | DKFZp686I04222 | MGC111370 | MSTP057 | PI6 | PTI | SPI3 | serpin peptidase inhibitor, clade B (ovalbumin), member 6 | - | HPRD | 8136380 |
| SERPINB8 | CAP2 | PI8 | serpin peptidase inhibitor, clade B (ovalbumin), member 8 | - | HPRD,BioGRID | 9402754 |
| TFPI | EPI | LACI | TFI | TFPI1 | tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) | - | HPRD,BioGRID | 3422166 |12787023 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG COMPLEMENT AND COAGULATION CASCADES | 69 | 49 | All SZGR 2.0 genes in this pathway |
| BIOCARTA AMI PATHWAY | 20 | 13 | All SZGR 2.0 genes in this pathway |
| BIOCARTA EXTRINSIC PATHWAY | 13 | 9 | All SZGR 2.0 genes in this pathway |
| BIOCARTA INTRINSIC PATHWAY | 23 | 15 | All SZGR 2.0 genes in this pathway |
| PID INTEGRIN2 PATHWAY | 29 | 18 | All SZGR 2.0 genes in this pathway |
| REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | 27 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | 11 | 8 | All SZGR 2.0 genes in this pathway |
| REACTOME COMMON PATHWAY | 14 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF PROTEINS | 518 | 242 | All SZGR 2.0 genes in this pathway |
| REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | 32 | 22 | All SZGR 2.0 genes in this pathway |
| REACTOME POST TRANSLATIONAL PROTEIN MODIFICATION | 188 | 116 | All SZGR 2.0 genes in this pathway |
| REACTOME INTRINSIC PATHWAY | 17 | 12 | All SZGR 2.0 genes in this pathway |
| REACTOME HEMOSTASIS | 466 | 331 | All SZGR 2.0 genes in this pathway |
| WONG ENDMETRIUM CANCER DN | 82 | 53 | All SZGR 2.0 genes in this pathway |
| BROWN MYELOID CELL DEVELOPMENT UP | 165 | 100 | All SZGR 2.0 genes in this pathway |
| WANG TARGETS OF MLL CBP FUSION UP | 44 | 26 | All SZGR 2.0 genes in this pathway |
| GUO HEX TARGETS DN | 65 | 36 | All SZGR 2.0 genes in this pathway |
| HSIAO LIVER SPECIFIC GENES | 244 | 154 | All SZGR 2.0 genes in this pathway |
| KUMAR TARGETS OF MLL AF9 FUSION | 405 | 264 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS LATE PROGENITOR | 544 | 307 | All SZGR 2.0 genes in this pathway |
| JOSEPH RESPONSE TO SODIUM BUTYRATE DN | 64 | 45 | All SZGR 2.0 genes in this pathway |
| KAAB HEART ATRIUM VS VENTRICLE UP | 249 | 170 | All SZGR 2.0 genes in this pathway |
| RAMASWAMY METASTASIS DN | 61 | 47 | All SZGR 2.0 genes in this pathway |
| FOSTER TOLERANT MACROPHAGE UP | 156 | 92 | All SZGR 2.0 genes in this pathway |
| COATES MACROPHAGE M1 VS M2 DN | 78 | 44 | All SZGR 2.0 genes in this pathway |
| ACEVEDO NORMAL TISSUE ADJACENT TO LIVER TUMOR DN | 354 | 216 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER NORMAL LIKE UP | 476 | 285 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL UP | 260 | 174 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL CULTURED VS FRESH UP | 425 | 298 | All SZGR 2.0 genes in this pathway |
| LABBE WNT3A TARGETS UP | 112 | 71 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER SURVIVAL UP | 185 | 112 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| CHANG CORE SERUM RESPONSE DN | 209 | 137 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS UP | 491 | 316 | All SZGR 2.0 genes in this pathway |
| CHEN METABOLIC SYNDROM NETWORK | 1210 | 725 | All SZGR 2.0 genes in this pathway |
| WINNEPENNINCKX MELANOMA METASTASIS DN | 46 | 26 | All SZGR 2.0 genes in this pathway |
| CAIRO LIVER DEVELOPMENT DN | 222 | 141 | All SZGR 2.0 genes in this pathway |
| CHYLA CBFA2T3 TARGETS UP | 387 | 225 | All SZGR 2.0 genes in this pathway |
| ZHOU CELL CYCLE GENES IN IR RESPONSE 6HR | 85 | 49 | All SZGR 2.0 genes in this pathway |
| ZHOU CELL CYCLE GENES IN IR RESPONSE 24HR | 128 | 73 | All SZGR 2.0 genes in this pathway |
| NABA ECM REGULATORS | 238 | 125 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME ASSOCIATED | 753 | 411 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME | 1028 | 559 | All SZGR 2.0 genes in this pathway |