Gene Page: FBLN2
Summary ?
| GeneID | 2199 |
| Symbol | FBLN2 |
| Synonyms | - |
| Description | fibulin 2 |
| Reference | MIM:135821|HGNC:HGNC:3601|Ensembl:ENSG00000163520|HPRD:00630|Vega:OTTHUMG00000155437 |
| Gene type | protein-coding |
| Map location | 3p25.1 |
| Pascal p-value | 0.815 |
| Sherlock p-value | 0.766 |
| Fetal beta | -2.424 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| GSMA_I | Genome scan meta-analysis | Psr: 0.006 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg23043139 | 3 | 13678918 | FBLN2 | 1.46E-4 | 0.345 | 0.031 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs17646919 | chr22 | 30400860 | FBLN2 | 2199 | 0.01 | trans |
Section II. Transcriptome annotation
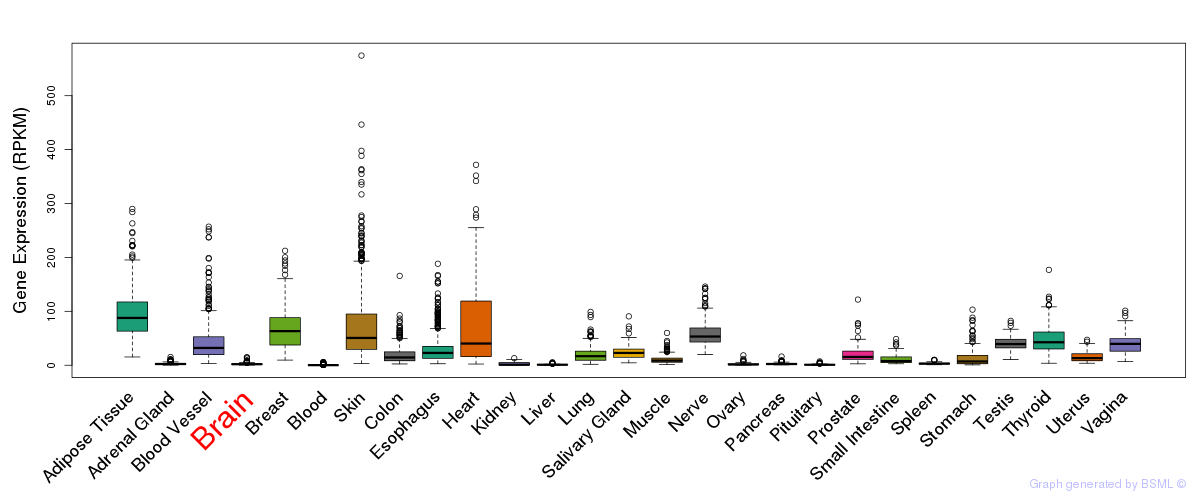
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SIGLEC10 | 0.87 | 0.79 |
| FCGR2A | 0.84 | 0.78 |
| LAPTM5 | 0.83 | 0.78 |
| ITGB2 | 0.83 | 0.77 |
| NCKAP1L | 0.82 | 0.75 |
| SIGLEC8 | 0.82 | 0.72 |
| LCP1 | 0.82 | 0.76 |
| HCK | 0.82 | 0.72 |
| TLR7 | 0.80 | 0.71 |
| CD53 | 0.80 | 0.77 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AC098691.2 | -0.27 | -0.36 |
| AF347015.18 | -0.24 | -0.27 |
| AC016705.1 | -0.23 | -0.21 |
| AC010300.1 | -0.22 | -0.30 |
| ANP32C | -0.21 | -0.20 |
| MT-CO2 | -0.21 | -0.27 |
| AF347015.21 | -0.21 | -0.24 |
| AC100783.1 | -0.21 | -0.22 |
| AF347015.26 | -0.21 | -0.22 |
| AC073957.1 | -0.21 | -0.21 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005509 | calcium ion binding | IEA | - | |
| GO:0005509 | calcium ion binding | TAS | 7806230 | |
| GO:0005201 | extracellular matrix structural constituent | TAS | 7806230 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005576 | extracellular region | IEA | - | |
| GO:0005578 | proteinaceous extracellular matrix | TAS | 7806230 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ACAN | AGC1 | AGCAN | CSPG1 | CSPGCP | MSK16 | SEDK | aggrecan | - | HPRD,BioGRID | 11038354 |
| BCAN | BEHAB | CSPG7 | MGC13038 | brevican | - | HPRD,BioGRID | 11038354 |
| COL18A1 | FLJ27325 | FLJ34914 | KNO | KNO1 | MGC74745 | collagen, type XVIII, alpha 1 | - | HPRD | 10544250 |
| COL4A1 | arresten | collagen, type IV, alpha 1 | - | HPRD,BioGRID | 7500359 |
| COL4A2 | DKFZp686I14213 | FLJ22259 | collagen, type IV, alpha 2 | - | HPRD,BioGRID | 7500359 |
| COL4A3 | - | collagen, type IV, alpha 3 (Goodpasture antigen) | - | HPRD,BioGRID | 7500359 |
| COL4A4 | CA44 | collagen, type IV, alpha 4 | - | HPRD,BioGRID | 7500359 |
| COL4A5 | ASLN | ATS | CA54 | MGC167109 | MGC42377 | collagen, type IV, alpha 5 | - | HPRD,BioGRID | 7500359 |
| COL4A6 | MGC88184 | collagen, type IV, alpha 6 | - | HPRD,BioGRID | 7500359 |
| ELN | FLJ38671 | FLJ43523 | SVAS | WBS | WS | elastin | - | HPRD,BioGRID | 10544250 |
| FBLN2 | - | fibulin 2 | - | HPRD,BioGRID | 9214621 |
| FBN1 | FBN | MASS | MFS1 | OCTD | SGS | WMS | fibrillin 1 | Fibrillin-1 interacts with Fibulin-2. | BIND | 8702639 |
| FBN1 | FBN | MASS | MFS1 | OCTD | SGS | WMS | fibrillin 1 | - | HPRD,BioGRID | 8702639 |9886271 |
| FN1 | CIG | DKFZp686F10164 | DKFZp686H0342 | DKFZp686I1370 | DKFZp686O13149 | ED-B | FINC | FN | FNZ | GFND | GFND2 | LETS | MSF | fibronectin 1 | - | HPRD | 10848816 |
| HSPG2 | PLC | PRCAN | SJA | SJS | SJS1 | heparan sulfate proteoglycan 2 | - | HPRD | 9431988 |
| HSPG2 | PLC | PRCAN | SJA | SJS | SJS1 | heparan sulfate proteoglycan 2 | Reconstituted Complex | BioGRID | 11493006 |
| LAMA1 | LAMA | laminin, alpha 1 | - | HPRD,BioGRID | 9006922 |10934193 |
| LAMA5 | KIAA1907 | laminin, alpha 5 | - | HPRD,BioGRID | 9006922 |
| LAMC2 | B2T | BM600 | CSF | EBR2 | EBR2A | LAMB2T | LAMNB2 | MGC138491 | MGC141938 | laminin, gamma 2 | - | HPRD | 11733994 |
| NID1 | NID | nidogen 1 | - | HPRD,BioGRID | 7500359 |11493006 |
| PRELP | MGC45323 | MST161 | MSTP161 | SLRR2A | proline/arginine-rich end leucine-rich repeat protein | - | HPRD,BioGRID | 11847210 |
| VCAN | CSPG2 | DKFZp686K06110 | ERVR | PG-M | WGN | WGN1 | versican | - | HPRD,BioGRID | 11038354 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| SCHUETZ BREAST CANCER DUCTAL INVASIVE UP | 351 | 230 | All SZGR 2.0 genes in this pathway |
| CASTELLANO NRAS TARGETS DN | 14 | 14 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 1 DN | 378 | 231 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 2B | 392 | 251 | All SZGR 2.0 genes in this pathway |
| GRUETZMANN PANCREATIC CANCER UP | 358 | 245 | All SZGR 2.0 genes in this pathway |
| SASAI RESISTANCE TO NEOPLASTIC TRANSFROMATION | 50 | 31 | All SZGR 2.0 genes in this pathway |
| KOYAMA SEMA3B TARGETS DN | 411 | 249 | All SZGR 2.0 genes in this pathway |
| GOTZMANN EPITHELIAL TO MESENCHYMAL TRANSITION UP | 69 | 55 | All SZGR 2.0 genes in this pathway |
| SUNG METASTASIS STROMA UP | 110 | 70 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER DENA UP | 60 | 40 | All SZGR 2.0 genes in this pathway |
| NADLER OBESITY UP | 61 | 34 | All SZGR 2.0 genes in this pathway |
| GUO HEX TARGETS UP | 81 | 54 | All SZGR 2.0 genes in this pathway |
| ZHAN MULTIPLE MYELOMA SPIKED | 22 | 13 | All SZGR 2.0 genes in this pathway |
| IGLESIAS E2F TARGETS UP | 151 | 103 | All SZGR 2.0 genes in this pathway |
| KAAB HEART ATRIUM VS VENTRICLE UP | 249 | 170 | All SZGR 2.0 genes in this pathway |
| YAMAZAKI TCEB3 TARGETS UP | 175 | 116 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS 7 | 51 | 34 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL AND PROGENITOR | 681 | 420 | All SZGR 2.0 genes in this pathway |
| RAMASWAMY METASTASIS UP | 66 | 43 | All SZGR 2.0 genes in this pathway |
| LEE CALORIE RESTRICTION MUSCLE DN | 51 | 28 | All SZGR 2.0 genes in this pathway |
| SANSOM APC TARGETS | 212 | 121 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE BY 4NQO | 38 | 24 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE DN | 195 | 135 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS UP | 673 | 430 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS UP | 602 | 364 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS DN | 591 | 366 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER LUMINAL B DN | 564 | 326 | All SZGR 2.0 genes in this pathway |
| MCGARVEY SILENCED BY METHYLATION IN COLON CANCER | 42 | 29 | All SZGR 2.0 genes in this pathway |
| ZHENG GLIOBLASTOMA PLASTICITY UP | 250 | 168 | All SZGR 2.0 genes in this pathway |
| LABBE WNT3A TARGETS UP | 112 | 71 | All SZGR 2.0 genes in this pathway |
| LABBE TGFB1 TARGETS UP | 102 | 64 | All SZGR 2.0 genes in this pathway |
| LABBE TARGETS OF TGFB1 AND WNT3A UP | 111 | 70 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS 60HR DN | 277 | 166 | All SZGR 2.0 genes in this pathway |
| CHEN METABOLIC SYNDROM NETWORK | 1210 | 725 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3 | 1069 | 729 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA CHEMOTAXIS DN | 457 | 302 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3K4ME2 | 491 | 319 | All SZGR 2.0 genes in this pathway |
| WONG ADULT TISSUE STEM MODULE | 721 | 492 | All SZGR 2.0 genes in this pathway |
| KARLSSON TGFB1 TARGETS UP | 127 | 78 | All SZGR 2.0 genes in this pathway |
| PLASARI TGFB1 TARGETS 1HR UP | 34 | 26 | All SZGR 2.0 genes in this pathway |
| DELACROIX RARG BOUND MEF | 367 | 231 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 2ND EGF PULSE ONLY | 1725 | 838 | All SZGR 2.0 genes in this pathway |
| NABA ECM GLYCOPROTEINS | 196 | 99 | All SZGR 2.0 genes in this pathway |
| NABA CORE MATRISOME | 275 | 148 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME | 1028 | 559 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-128 | 355 | 362 | 1A,m8 | hsa-miR-128a | UCACAGUGAACCGGUCUCUUUU |
| hsa-miR-128b | UCACAGUGAACCGGUCUCUUUC | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.