Gene Page: SYNE1
Summary ?
| GeneID | 23345 |
| Symbol | SYNE1 |
| Synonyms | 8B|ARCA1|C6orf98|CPG2|EDMD4|MYNE1|Nesp1|SCAR8|dJ45H2.2 |
| Description | spectrin repeat containing, nuclear envelope 1 |
| Reference | MIM:608441|HGNC:HGNC:17089|Ensembl:ENSG00000131018|HPRD:09762|Vega:OTTHUMG00000015841 |
| Gene type | protein-coding |
| Map location | 6q25 |
| Pascal p-value | 0.003 |
| Sherlock p-value | 0.419 |
| TADA p-value | 0.714 |
| Fetal beta | -1.223 |
| Support | G2Cdb.humanPSD G2Cdb.humanPSP CompositeSet Darnell FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWAScat | Genome-wide Association Studies | This data set includes 560 SNPs associated with schizophrenia. A total of 486 genes were mapped to these SNPs within 50kb. | |
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DNM:Fromer_2014 | Whole Exome Sequencing analysis | This study reported a WES study of 623 schizophrenia trios, reporting DNMs using genomic DNA. | |
| DNM:Guipponi_2014 | Whole Exome Sequencing analysis | 49 DNMs were identified by comparing the exome of 53 individuals with sporadic SCZ and of their non-affected parents | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.6157 |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| SYNE1 | chr6 | 152545708 | T | G | NM_033071 NM_182961 | p.7077E>A p.7148E>A | missense missense | Schizophrenia | DNM:Fromer_2014 | ||
| SYNE1 | A | T | NM_033071 | p.Q6633L | missense | 1 | 0.13 | Schizophrenia | DNM:Guipponi_2014 |
Section II. Transcriptome annotation
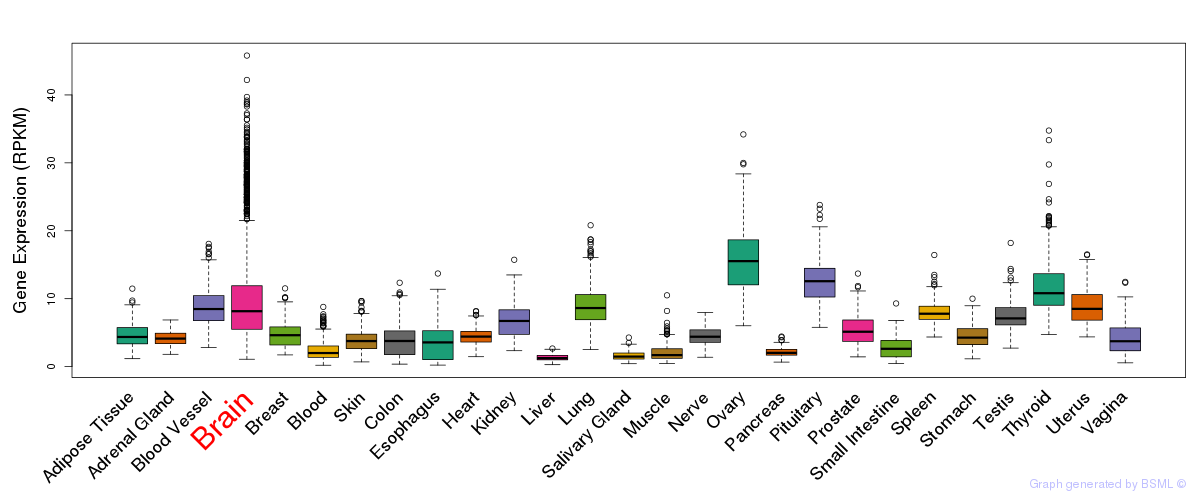
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003779 | actin binding | IDA | 12408964 | |
| GO:0003779 | actin binding | IEA | - | |
| GO:0005515 | protein binding | IEA | - | |
| GO:0005521 | lamin binding | IPI | 11801724 | |
| GO:0005198 | structural molecule activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0008219 | cell death | IEA | - | |
| GO:0007030 | Golgi organization | IDA | 12808039 | |
| GO:0006997 | nucleus organization | NAS | 11792814 | |
| GO:0042692 | muscle cell differentiation | IDA | 11792814 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0045211 | postsynaptic membrane | IDA | Synap, Neurotransmitter (GO term level: 5) | 10878022 |
| GO:0005794 | Golgi apparatus | IDA | 12808039 | |
| GO:0005856 | cytoskeleton | IEA | - | |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005635 | nuclear envelope | IDA | 11792814 | |
| GO:0005635 | nuclear envelope | IEA | - | |
| GO:0005640 | nuclear outer membrane | IEA | - | |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0016021 | integral to membrane | IDA | 11792814 | |
| GO:0016021 | integral to membrane | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| REACTOME MEIOSIS | 116 | 81 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE | 421 | 253 | All SZGR 2.0 genes in this pathway |
| REACTOME CHROMOSOME MAINTENANCE | 122 | 80 | All SZGR 2.0 genes in this pathway |
| REACTOME MEIOTIC SYNAPSIS | 73 | 57 | All SZGR 2.0 genes in this pathway |
| WINTER HYPOXIA DN | 52 | 30 | All SZGR 2.0 genes in this pathway |
| PARENT MTOR SIGNALING UP | 567 | 375 | All SZGR 2.0 genes in this pathway |
| HOLLMANN APOPTOSIS VIA CD40 UP | 201 | 125 | All SZGR 2.0 genes in this pathway |
| THUM SYSTOLIC HEART FAILURE DN | 244 | 147 | All SZGR 2.0 genes in this pathway |
| DEURIG T CELL PROLYMPHOCYTIC LEUKEMIA DN | 320 | 184 | All SZGR 2.0 genes in this pathway |
| ODONNELL TFRC TARGETS UP | 456 | 228 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC3 TARGETS DN | 536 | 332 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS 8HR UP | 164 | 122 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| DELYS THYROID CANCER DN | 232 | 154 | All SZGR 2.0 genes in this pathway |
| CHEBOTAEV GR TARGETS DN | 120 | 73 | All SZGR 2.0 genes in this pathway |
| LUI THYROID CANCER CLUSTER 2 | 42 | 31 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS UP | 1037 | 673 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 2 UP | 256 | 159 | All SZGR 2.0 genes in this pathway |
| KENNY CTNNB1 TARGETS UP | 50 | 30 | All SZGR 2.0 genes in this pathway |
| NAKAJIMA EOSINOPHIL | 30 | 20 | All SZGR 2.0 genes in this pathway |
| WANG TARGETS OF MLL CBP FUSION UP | 44 | 26 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 14HR DN | 298 | 200 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 20HR DN | 101 | 70 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RESPONSE TO TRABECTEDIN DN | 271 | 175 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER NORMAL LIKE UP | 476 | 285 | All SZGR 2.0 genes in this pathway |
| BONOME OVARIAN CANCER SURVIVAL SUBOPTIMAL DEBULKING | 510 | 309 | All SZGR 2.0 genes in this pathway |
| SHEDDEN LUNG CANCER GOOD SURVIVAL A4 | 196 | 124 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS 60HR UP | 293 | 203 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION DN | 1080 | 713 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-15/16/195/424/497 | 362 | 368 | m8 | hsa-miR-15abrain | UAGCAGCACAUAAUGGUUUGUG |
| hsa-miR-16brain | UAGCAGCACGUAAAUAUUGGCG | ||||
| hsa-miR-15bbrain | UAGCAGCACAUCAUGGUUUACA | ||||
| hsa-miR-195SZ | UAGCAGCACAGAAAUAUUGGC | ||||
| hsa-miR-424 | CAGCAGCAAUUCAUGUUUUGAA | ||||
| hsa-miR-497 | CAGCAGCACACUGUGGUUUGU | ||||
| miR-181 | 644 | 650 | m8 | hsa-miR-181abrain | AACAUUCAACGCUGUCGGUGAGU |
| hsa-miR-181bSZ | AACAUUCAUUGCUGUCGGUGGG | ||||
| hsa-miR-181cbrain | AACAUUCAACCUGUCGGUGAGU | ||||
| hsa-miR-181dbrain | AACAUUCAUUGUUGUCGGUGGGUU | ||||
| miR-23 | 631 | 637 | 1A | hsa-miR-23abrain | AUCACAUUGCCAGGGAUUUCC |
| hsa-miR-23bbrain | AUCACAUUGCCAGGGAUUACC | ||||
| miR-29 | 359 | 365 | m8 | hsa-miR-29aSZ | UAGCACCAUCUGAAAUCGGUU |
| hsa-miR-29bSZ | UAGCACCAUUUGAAAUCAGUGUU | ||||
| hsa-miR-29cSZ | UAGCACCAUUUGAAAUCGGU | ||||
| miR-369-3p | 729 | 735 | 1A | hsa-miR-369-3p | AAUAAUACAUGGUUGAUCUUU |
| miR-374 | 729 | 735 | m8 | hsa-miR-374 | UUAUAAUACAACCUGAUAAGUG |
| miR-493-5p | 679 | 685 | m8 | hsa-miR-493-5p | UUGUACAUGGUAGGCUUUCAUU |
| miR-543 | 645 | 652 | 1A,m8 | hsa-miR-543 | AAACAUUCGCGGUGCACUUCU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.