Gene Page: FXN
Summary ?
| GeneID | 2395 |
| Symbol | FXN |
| Synonyms | CyaY|FA|FARR|FRDA|X25 |
| Description | frataxin |
| Reference | MIM:606829|HGNC:HGNC:3951|Ensembl:ENSG00000165060|HPRD:06013| |
| Gene type | protein-coding |
| Map location | 9q21.11 |
| Pascal p-value | 0.041 |
| Sherlock p-value | 0.15 |
| DMG | 1 (# studies) |
| eGene | Anterior cingulate cortex BA24 Nucleus accumbens basal ganglia Putamen basal ganglia Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWAScat | Genome-wide Association Studies | This data set includes 560 SNPs associated with schizophrenia. A total of 486 genes were mapped to these SNPs within 50kb. | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg23667933 | 9 | 71650719 | FXN | 1.09E-9 | -0.018 | 1.23E-6 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs10751722 | chr1 | 27543522 | FXN | 2395 | 0.09 | trans | ||
| rs2289256 | chr4 | 187089132 | FXN | 2395 | 0.13 | trans | ||
| rs7858407 | 9 | 71686520 | FXN | ENSG00000165060.7 | 3.243E-6 | 0.05 | 36345 | gtex_brain_ba24 |
| rs3793459 | 9 | 71675211 | FXN | ENSG00000165060.7 | 4.705E-7 | 0 | 25036 | gtex_brain_putamen_basal |
| rs3793460 | 9 | 71675220 | FXN | ENSG00000165060.7 | 3.207E-6 | 0 | 25045 | gtex_brain_putamen_basal |
| rs9314854 | 9 | 71686181 | FXN | ENSG00000165060.7 | 4.476E-7 | 0 | 36006 | gtex_brain_putamen_basal |
| rs7870295 | 9 | 71686476 | FXN | ENSG00000165060.7 | 2.498E-7 | 0 | 36301 | gtex_brain_putamen_basal |
| rs7858407 | 9 | 71686520 | FXN | ENSG00000165060.7 | 5.895E-7 | 0 | 36345 | gtex_brain_putamen_basal |
| rs7861997 | 9 | 71686557 | FXN | ENSG00000165060.7 | 2.348E-7 | 0 | 36382 | gtex_brain_putamen_basal |
| rs7858657 | 9 | 71686702 | FXN | ENSG00000165060.7 | 9.006E-7 | 0 | 36527 | gtex_brain_putamen_basal |
| rs7870727 | 9 | 71686866 | FXN | ENSG00000165060.7 | 9.006E-7 | 0 | 36691 | gtex_brain_putamen_basal |
| rs1411676 | 9 | 71686918 | FXN | ENSG00000165060.7 | 9.006E-7 | 0 | 36743 | gtex_brain_putamen_basal |
| rs1411675 | 9 | 71687052 | FXN | ENSG00000165060.7 | 9.006E-7 | 0 | 36877 | gtex_brain_putamen_basal |
| rs3066313 | 9 | 71687400 | FXN | ENSG00000165060.7 | 9.006E-7 | 0 | 37225 | gtex_brain_putamen_basal |
| rs7025834 | 9 | 71687440 | FXN | ENSG00000165060.7 | 2.348E-7 | 0 | 37265 | gtex_brain_putamen_basal |
| rs529317691 | 9 | 71687755 | FXN | ENSG00000165060.7 | 2.348E-7 | 0 | 37580 | gtex_brain_putamen_basal |
| rs10890 | 9 | 71687924 | FXN | ENSG00000165060.7 | 9.006E-7 | 0 | 37749 | gtex_brain_putamen_basal |
| rs4745577 | 9 | 71688101 | FXN | ENSG00000165060.7 | 9.006E-7 | 0 | 37926 | gtex_brain_putamen_basal |
| rs4744806 | 9 | 71688218 | FXN | ENSG00000165060.7 | 9.006E-7 | 0 | 38043 | gtex_brain_putamen_basal |
| rs4744807 | 9 | 71688300 | FXN | ENSG00000165060.7 | 2.348E-7 | 0 | 38125 | gtex_brain_putamen_basal |
| rs4744808 | 9 | 71688311 | FXN | ENSG00000165060.7 | 9.006E-7 | 0 | 38136 | gtex_brain_putamen_basal |
| rs4744810 | 9 | 71689004 | FXN | ENSG00000165060.7 | 3.1E-6 | 0 | 38829 | gtex_brain_putamen_basal |
| rs2871218 | 9 | 71689045 | FXN | ENSG00000165060.7 | 4.827E-7 | 0 | 38870 | gtex_brain_putamen_basal |
| rs386361262 | 9 | 71689116 | FXN | ENSG00000165060.7 | 8.993E-7 | 0 | 38941 | gtex_brain_putamen_basal |
| rs1984004 | 9 | 71689375 | FXN | ENSG00000165060.7 | 9.196E-7 | 0 | 39200 | gtex_brain_putamen_basal |
| rs1888334 | 9 | 71689415 | FXN | ENSG00000165060.7 | 2.279E-7 | 0 | 39240 | gtex_brain_putamen_basal |
| rs200175861 | 9 | 71689447 | FXN | ENSG00000165060.7 | 2.434E-6 | 0 | 39272 | gtex_brain_putamen_basal |
| rs139068140 | 9 | 71689448 | FXN | ENSG00000165060.7 | 9.093E-7 | 0 | 39273 | gtex_brain_putamen_basal |
| rs1984003 | 9 | 71689465 | FXN | ENSG00000165060.7 | 1.482E-6 | 0 | 39290 | gtex_brain_putamen_basal |
| rs1984002 | 9 | 71689633 | FXN | ENSG00000165060.7 | 8.779E-7 | 0 | 39458 | gtex_brain_putamen_basal |
| rs11432103 | 9 | 71689659 | FXN | ENSG00000165060.7 | 5.944E-7 | 0 | 39484 | gtex_brain_putamen_basal |
| rs4745580 | 9 | 71689756 | FXN | ENSG00000165060.7 | 2.286E-7 | 0 | 39581 | gtex_brain_putamen_basal |
| rs4745581 | 9 | 71689880 | FXN | ENSG00000165060.7 | 8.813E-7 | 0 | 39705 | gtex_brain_putamen_basal |
| rs4745582 | 9 | 71690144 | FXN | ENSG00000165060.7 | 8.516E-7 | 0 | 39969 | gtex_brain_putamen_basal |
| rs150186989 | 9 | 71690145 | FXN | ENSG00000165060.7 | 1.273E-6 | 0 | 39970 | gtex_brain_putamen_basal |
| rs4745583 | 9 | 71690182 | FXN | ENSG00000165060.7 | 3.842E-7 | 0 | 40007 | gtex_brain_putamen_basal |
| rs7865854 | 9 | 71690246 | FXN | ENSG00000165060.7 | 2.803E-7 | 0 | 40071 | gtex_brain_putamen_basal |
| rs7866067 | 9 | 71690370 | FXN | ENSG00000165060.7 | 2.869E-7 | 0 | 40195 | gtex_brain_putamen_basal |
| rs7874989 | 9 | 71690501 | FXN | ENSG00000165060.7 | 3.236E-7 | 0 | 40326 | gtex_brain_putamen_basal |
| rs7875564 | 9 | 71690963 | FXN | ENSG00000165060.7 | 9.252E-7 | 0 | 40788 | gtex_brain_putamen_basal |
| rs7875659 | 9 | 71690974 | FXN | ENSG00000165060.7 | 9.004E-7 | 0 | 40799 | gtex_brain_putamen_basal |
| rs7858249 | 9 | 71691030 | FXN | ENSG00000165060.7 | 9.002E-7 | 0 | 40855 | gtex_brain_putamen_basal |
| rs10781379 | 9 | 71691239 | FXN | ENSG00000165060.7 | 9.006E-7 | 0 | 41064 | gtex_brain_putamen_basal |
| rs7874105 | 9 | 71692080 | FXN | ENSG00000165060.7 | 8.011E-7 | 0 | 41905 | gtex_brain_putamen_basal |
| rs11145063 | 9 | 71692137 | FXN | ENSG00000165060.7 | 2.372E-7 | 0 | 41962 | gtex_brain_putamen_basal |
| rs10869821 | 9 | 71692385 | FXN | ENSG00000165060.7 | 1.129E-6 | 0 | 42210 | gtex_brain_putamen_basal |
| rs11145068 | 9 | 71692811 | FXN | ENSG00000165060.7 | 9.235E-7 | 0 | 42636 | gtex_brain_putamen_basal |
| rs11145069 | 9 | 71692824 | FXN | ENSG00000165060.7 | 1.665E-6 | 0 | 42649 | gtex_brain_putamen_basal |
| rs35104955 | 9 | 71692846 | FXN | ENSG00000165060.7 | 2.352E-7 | 0 | 42671 | gtex_brain_putamen_basal |
| rs11145070 | 9 | 71692862 | FXN | ENSG00000165060.7 | 1.279E-6 | 0 | 42687 | gtex_brain_putamen_basal |
| rs11145072 | 9 | 71692944 | FXN | ENSG00000165060.7 | 1.299E-6 | 0 | 42769 | gtex_brain_putamen_basal |
| rs10869823 | 9 | 71693079 | FXN | ENSG00000165060.7 | 1.347E-6 | 0 | 42904 | gtex_brain_putamen_basal |
| rs7039631 | 9 | 71693292 | FXN | ENSG00000165060.7 | 1.417E-6 | 0 | 43117 | gtex_brain_putamen_basal |
| rs7024295 | 9 | 71693481 | FXN | ENSG00000165060.7 | 1.483E-6 | 0 | 43306 | gtex_brain_putamen_basal |
| rs11301594 | 9 | 71693601 | FXN | ENSG00000165060.7 | 1.543E-6 | 0 | 43426 | gtex_brain_putamen_basal |
| rs57561255 | 9 | 71693609 | FXN | ENSG00000165060.7 | 1.53E-6 | 0 | 43434 | gtex_brain_putamen_basal |
| rs1045632 | 9 | 71693777 | FXN | ENSG00000165060.7 | 1.596E-6 | 0 | 43602 | gtex_brain_putamen_basal |
| rs4745585 | 9 | 71694084 | FXN | ENSG00000165060.7 | 2.453E-6 | 0 | 43909 | gtex_brain_putamen_basal |
| rs4744811 | 9 | 71694139 | FXN | ENSG00000165060.7 | 2.348E-7 | 0 | 43964 | gtex_brain_putamen_basal |
| rs11145079 | 9 | 71694181 | FXN | ENSG00000165060.7 | 2.703E-6 | 0 | 44006 | gtex_brain_putamen_basal |
| rs7859021 | 9 | 71694259 | FXN | ENSG00000165060.7 | 1.677E-6 | 0 | 44084 | gtex_brain_putamen_basal |
| rs7028835 | 9 | 71694592 | FXN | ENSG00000165060.7 | 1.618E-6 | 0 | 44417 | gtex_brain_putamen_basal |
| rs7035156 | 9 | 71694719 | FXN | ENSG00000165060.7 | 1.631E-6 | 0 | 44544 | gtex_brain_putamen_basal |
| rs10869824 | 9 | 71694927 | FXN | ENSG00000165060.7 | 2.02E-6 | 0 | 44752 | gtex_brain_putamen_basal |
| rs56078943 | 9 | 71695574 | FXN | ENSG00000165060.7 | 1.629E-6 | 0 | 45399 | gtex_brain_putamen_basal |
| rs56102961 | 9 | 71695688 | FXN | ENSG00000165060.7 | 1.864E-6 | 0 | 45513 | gtex_brain_putamen_basal |
| rs12348477 | 9 | 71695867 | FXN | ENSG00000165060.7 | 1.729E-6 | 0 | 45692 | gtex_brain_putamen_basal |
| rs7041324 | 9 | 71695925 | FXN | ENSG00000165060.7 | 1.677E-6 | 0 | 45750 | gtex_brain_putamen_basal |
| rs6560545 | 9 | 71696140 | FXN | ENSG00000165060.7 | 1.679E-6 | 0 | 45965 | gtex_brain_putamen_basal |
| rs6560546 | 9 | 71696411 | FXN | ENSG00000165060.7 | 1.679E-6 | 0 | 46236 | gtex_brain_putamen_basal |
| rs6560547 | 9 | 71696685 | FXN | ENSG00000165060.7 | 2.498E-7 | 0 | 46510 | gtex_brain_putamen_basal |
| rs6560550 | 9 | 71697916 | FXN | ENSG00000165060.7 | 2.345E-7 | 0 | 47741 | gtex_brain_putamen_basal |
| rs6560551 | 9 | 71698202 | FXN | ENSG00000165060.7 | 2.498E-7 | 0 | 48027 | gtex_brain_putamen_basal |
| rs11145105 | 9 | 71698978 | FXN | ENSG00000165060.7 | 2.493E-7 | 0 | 48803 | gtex_brain_putamen_basal |
| rs10747003 | 9 | 71702098 | FXN | ENSG00000165060.7 | 1.538E-6 | 0 | 51923 | gtex_brain_putamen_basal |
| rs10116434 | 9 | 71702821 | FXN | ENSG00000165060.7 | 1.294E-6 | 0 | 52646 | gtex_brain_putamen_basal |
| rs11145124 | 9 | 71702999 | FXN | ENSG00000165060.7 | 1.018E-6 | 0 | 52824 | gtex_brain_putamen_basal |
| rs62567097 | 9 | 71730265 | FXN | ENSG00000165060.7 | 3.449E-6 | 0 | 80090 | gtex_brain_putamen_basal |
| rs11145326 | 9 | 71733142 | FXN | ENSG00000165060.7 | 3.792E-6 | 0 | 82967 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
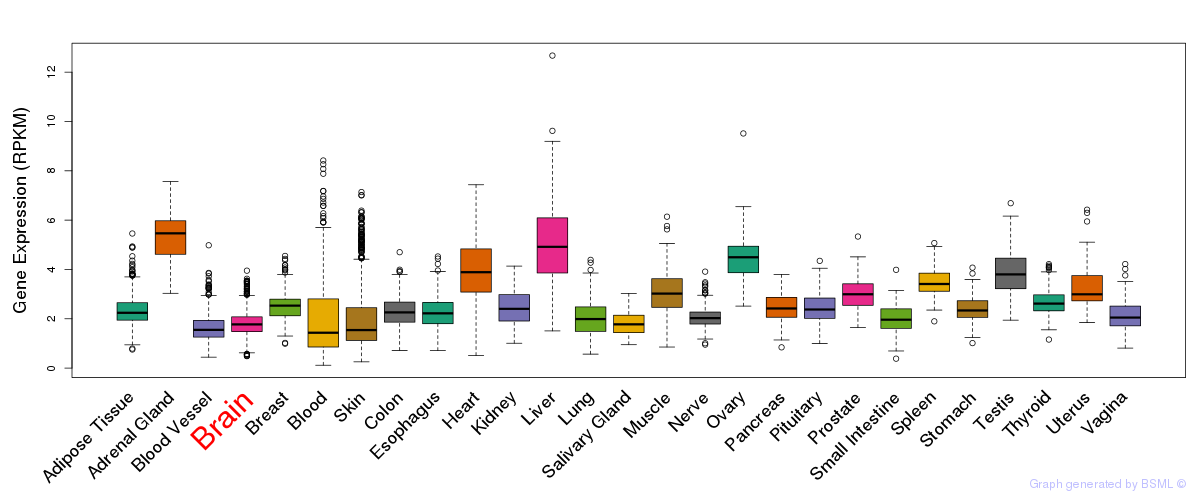
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005381 | iron ion transmembrane transporter activity | TAS | 9326946 | |
| GO:0004428 | inositol or phosphatidylinositol kinase activity | TAS | 8841185 | |
| GO:0009055 | electron carrier activity | TAS | 9326946 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007268 | synaptic transmission | TAS | neuron, Synap, Neurotransmitter (GO term level: 6) | 8841185 |
| GO:0006879 | cellular iron ion homeostasis | IEA | - | |
| GO:0006879 | cellular iron ion homeostasis | TAS | 9180083 | |
| GO:0016192 | vesicle-mediated transport | TAS | 8841185 | |
| GO:0055114 | oxidation reduction | TAS | 9180083 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005739 | mitochondrion | IEA | - | |
| GO:0005739 | mitochondrion | TAS | 9241270 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PID HIF2PATHWAY | 34 | 29 | All SZGR 2.0 genes in this pathway |
| REACTOME MITOCHONDRIAL PROTEIN IMPORT | 58 | 24 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF PROTEINS | 518 | 242 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS 12HR DN | 209 | 122 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN DN | 770 | 415 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER MYC TARGETS AND SERUM RESPONSE UP | 47 | 32 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| PUJANA CHEK2 PCC NETWORK | 779 | 480 | All SZGR 2.0 genes in this pathway |
| GARCIA TARGETS OF FLI1 AND DAX1 DN | 176 | 104 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC TARGETS WITH EBOX | 230 | 156 | All SZGR 2.0 genes in this pathway |
| MANALO HYPOXIA DN | 289 | 166 | All SZGR 2.0 genes in this pathway |
| FERNANDEZ BOUND BY MYC | 182 | 116 | All SZGR 2.0 genes in this pathway |
| SCHUHMACHER MYC TARGETS UP | 80 | 57 | All SZGR 2.0 genes in this pathway |
| GUO HEX TARGETS DN | 65 | 36 | All SZGR 2.0 genes in this pathway |
| HOEGERKORP CD44 TARGETS TEMPORAL DN | 25 | 16 | All SZGR 2.0 genes in this pathway |
| RAMASWAMY METASTASIS DN | 61 | 47 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 UP | 428 | 266 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS 6 | 189 | 112 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR DN | 911 | 527 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR DN | 1011 | 592 | All SZGR 2.0 genes in this pathway |
| SANSOM WNT PATHWAY REQUIRE MYC | 58 | 43 | All SZGR 2.0 genes in this pathway |
| STEARMAN LUNG CANCER EARLY VS LATE UP | 125 | 89 | All SZGR 2.0 genes in this pathway |
| MOOTHA HUMAN MITODB 6 2002 | 429 | 260 | All SZGR 2.0 genes in this pathway |
| MOOTHA MITOCHONDRIA | 447 | 277 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA CLASSES UP | 605 | 377 | All SZGR 2.0 genes in this pathway |
| KYNG WERNER SYNDROM AND NORMAL AGING DN | 225 | 124 | All SZGR 2.0 genes in this pathway |
| DANG MYC TARGETS UP | 143 | 100 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| DELACROIX RAR BOUND ES | 462 | 273 | All SZGR 2.0 genes in this pathway |
| PHONG TNF RESPONSE NOT VIA P38 | 337 | 236 | All SZGR 2.0 genes in this pathway |