Gene Page: GHSR
Summary ?
| GeneID | 2693 |
| Symbol | GHSR |
| Synonyms | GHDP |
| Description | growth hormone secretagogue receptor |
| Reference | MIM:601898|HGNC:HGNC:4267|Ensembl:ENSG00000121853|HPRD:03541|Vega:OTTHUMG00000156946 |
| Gene type | protein-coding |
| Map location | 3q26.31 |
| Pascal p-value | 0.627 |
| Fetal beta | 0.173 |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 2 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs10994209 | chr10 | 61877705 | GHSR | 2693 | 0.16 | trans |
Section II. Transcriptome annotation
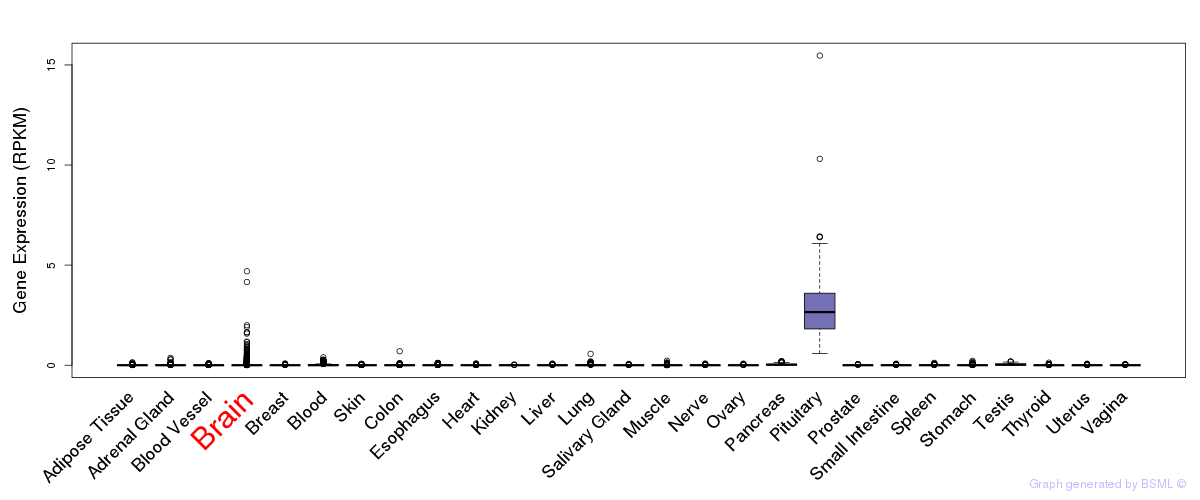
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| CABP1 | 0.82 | 0.68 |
| CCDC64B | 0.76 | 0.65 |
| GLS2 | 0.75 | 0.68 |
| ANXA11 | 0.74 | 0.73 |
| ABTB1 | 0.74 | 0.68 |
| TECR | 0.74 | 0.62 |
| ISCU | 0.74 | 0.66 |
| ACTR1B | 0.74 | 0.60 |
| PRSS16 | 0.74 | 0.64 |
| P2RX6 | 0.73 | 0.69 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| PKN1 | -0.44 | -0.51 |
| PDE9A | -0.42 | -0.53 |
| NKIRAS2 | -0.42 | -0.36 |
| SH2B2 | -0.41 | -0.54 |
| TUBB2B | -0.40 | -0.54 |
| SH3BP2 | -0.40 | -0.52 |
| KIAA1949 | -0.40 | -0.48 |
| FADS2 | -0.39 | -0.41 |
| MARCKSL1 | -0.39 | -0.44 |
| YBX1 | -0.39 | -0.48 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0001616 | growth hormone secretagogue receptor activity | IDA | 8688086 | |
| GO:0004872 | receptor activity | IEA | - | |
| GO:0016520 | growth hormone-releasing hormone receptor activity | IDA | 8688086 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0051963 | regulation of synaptogenesis | IEA | Synap (GO term level: 9) | - |
| GO:0030252 | growth hormone secretion | TAS | 16511605 | |
| GO:0007186 | G-protein coupled receptor protein signaling pathway | IDA | 8688086 | |
| GO:0007165 | signal transduction | IEA | - | |
| GO:0008343 | adult feeding behavior | ISS | - | |
| GO:0009755 | hormone-mediated signaling | IDA | 8688086 | |
| GO:0008154 | actin polymerization or depolymerization | IDA | 15232612 | |
| GO:0032100 | positive regulation of appetite | ISS | - | |
| GO:0040018 | positive regulation of multicellular organism growth | IMP | 16511605 | |
| GO:0032691 | negative regulation of interleukin-1 beta production | IDA | 15232612 | |
| GO:0042536 | negative regulation of tumor necrosis factor biosynthetic process | IDA | 15232612 | |
| GO:0050728 | negative regulation of inflammatory response | IDA | 15232612 | |
| GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway | IEA | - | |
| GO:0046697 | decidualization | IDA | 17494105 | |
| GO:0045409 | negative regulation of interleukin-6 biosynthetic process | IDA | 15232612 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0043005 | neuron projection | IDA | neuron, axon, neurite, dendrite (GO term level: 5) | 12890514 |
| GO:0016021 | integral to membrane | IEA | - | |
| GO:0009986 | cell surface | IDA | 8688086 | |
| GO:0005886 | plasma membrane | IEA | - | |
| GO:0045121 | membrane raft | IDA | 15232612 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG NEUROACTIVE LIGAND RECEPTOR INTERACTION | 272 | 195 | All SZGR 2.0 genes in this pathway |
| REACTOME GASTRIN CREB SIGNALLING PATHWAY VIA PKC AND MAPK | 205 | 136 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY GPCR | 920 | 449 | All SZGR 2.0 genes in this pathway |
| REACTOME PEPTIDE LIGAND BINDING RECEPTORS | 188 | 108 | All SZGR 2.0 genes in this pathway |
| REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | 305 | 177 | All SZGR 2.0 genes in this pathway |
| REACTOME G ALPHA Q SIGNALLING EVENTS | 184 | 116 | All SZGR 2.0 genes in this pathway |
| REACTOME GPCR DOWNSTREAM SIGNALING | 805 | 368 | All SZGR 2.0 genes in this pathway |
| REACTOME GPCR LIGAND BINDING | 408 | 246 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE FIMA DN | 284 | 156 | All SZGR 2.0 genes in this pathway |
| BENPORATH SUZ12 TARGETS | 1038 | 678 | All SZGR 2.0 genes in this pathway |
| BENPORATH EED TARGETS | 1062 | 725 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| BENPORATH PRC2 TARGETS | 652 | 441 | All SZGR 2.0 genes in this pathway |
| BANDRES RESPONSE TO CARMUSTIN MGMT 48HR DN | 161 | 105 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3 | 1069 | 729 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3K4ME2 AND H3K27ME3 | 349 | 234 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN NPC HCP WITH H3K27ME3 | 341 | 243 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MEF HCP WITH H3K27ME3 | 590 | 403 | All SZGR 2.0 genes in this pathway |
| ZWANG CLASS 1 TRANSIENTLY INDUCED BY EGF | 516 | 308 | All SZGR 2.0 genes in this pathway |