Gene Page: AMPH
Summary ?
| GeneID | 273 |
| Symbol | AMPH |
| Synonyms | AMPH1 |
| Description | amphiphysin |
| Reference | MIM:600418|HGNC:HGNC:471|Ensembl:ENSG00000078053|HPRD:02687|Vega:OTTHUMG00000023725 |
| Gene type | protein-coding |
| Map location | 7p14-p13 |
| Pascal p-value | 0.25 |
| Sherlock p-value | 0.345 |
| Fetal beta | -0.743 |
| eGene | Myers' cis & trans |
| Support | EXOCYTOSIS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS G2Cdb.human_clathrin G2Cdb.human_Synaptosome CompositeSet Darnell FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 4 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.252 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs7132043 | chr12 | 80968399 | AMPH | 273 | 0.11 | trans |
Section II. Transcriptome annotation
General gene expression (GTEx)
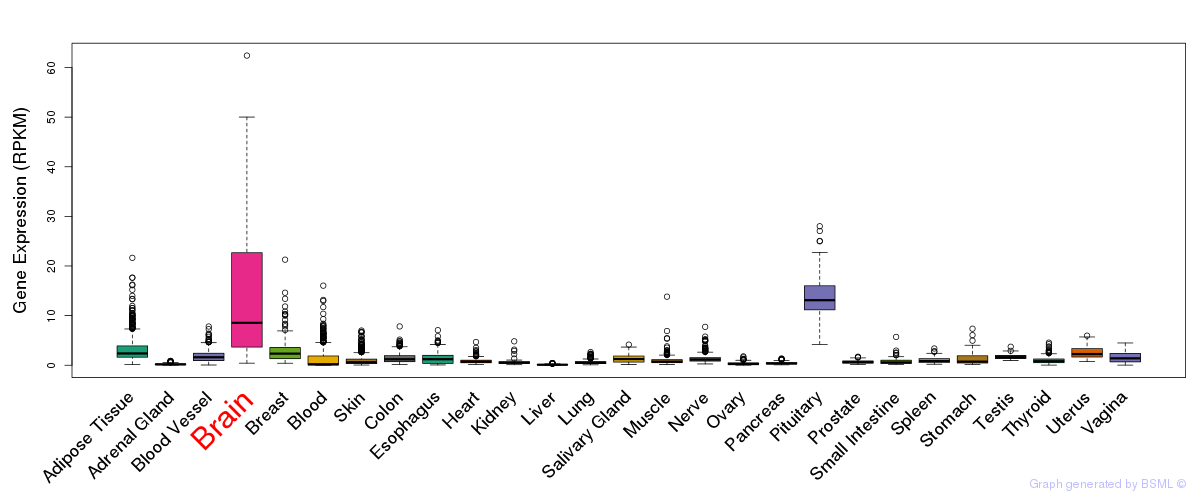
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005515 | protein binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007268 | synaptic transmission | TAS | neuron, Synap, Neurotransmitter (GO term level: 6) | 9182667 |
| GO:0048488 | synaptic vesicle endocytosis | IEA | axon, Synap, Neurotransmitter (GO term level: 7) | - |
| GO:0007612 | learning | IEA | - | |
| GO:0006897 | endocytosis | TAS | 9182667 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0008021 | synaptic vesicle | IEA | Synap, Neurotransmitter (GO term level: 12) | - |
| GO:0008021 | synaptic vesicle | TAS | Synap, Neurotransmitter (GO term level: 12) | 1628617 |
| GO:0045202 | synapse | IEA | neuron, Synap, Neurotransmitter, Glial (GO term level: 2) | - |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0016020 | membrane | IEA | - | |
| GO:0015629 | actin cytoskeleton | TAS | 9182667 | |
| GO:0030054 | cell junction | IEA | - | |
| GO:0031410 | cytoplasmic vesicle | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| AP1B1 | ADTB1 | AP105A | BAM22 | CLAPB2 | adaptor-related protein complex 1, beta 1 subunit | Reconstituted Complex | BioGRID | 9280305 |
| AP2A2 | ADTAB | CLAPA2 | HIP9 | HYPJ | adaptor-related protein complex 2, alpha 2 subunit | Reconstituted Complex | BioGRID | 9280305 |
| BIN1 | AMPH2 | AMPHL | DKFZp547F068 | MGC10367 | SH3P9 | bridging integrator 1 | - | HPRD,BioGRID | 9348539 |
| CABIN1 | CAIN | KIAA0330 | PPP3IN | calcineurin binding protein 1 | - | HPRD,BioGRID | 10931822 |
| CDK5R1 | CDK5P35 | CDK5R | MGC33831 | NCK5A | p23 | p25 | p35 | p35nck5a | cyclin-dependent kinase 5, regulatory subunit 1 (p35) | Amphiphysin 1 interacts with p35. This interaction was modeled on a demonstrated interaction between human amphiphysin 1 and rat p35. | BIND | 11113134 |
| CDK5R1 | CDK5P35 | CDK5R | MGC33831 | NCK5A | p23 | p25 | p35 | p35nck5a | cyclin-dependent kinase 5, regulatory subunit 1 (p35) | - | HPRD,BioGRID | 11113134 |
| CLTC | CHC | CHC17 | CLH-17 | CLTCL2 | Hc | KIAA0034 | clathrin, heavy chain (Hc) | - | HPRD | 9195986 |9280305 |9603201 |
| CLTC | CHC | CHC17 | CLH-17 | CLTCL2 | Hc | KIAA0034 | clathrin, heavy chain (Hc) | in vitro in vivo Reconstituted Complex | BioGRID | 9195986 |9603201 |
| DNM1 | DNM | dynamin 1 | Affinity Capture-Western Reconstituted Complex | BioGRID | 9148966 |9280305 |9341169 |9348539 |11877424 |
| DNM1 | DNM | dynamin 1 | - | HPRD | 9148966 |9348539 |9694653 |11877424|10899172 |
| PLD1 | - | phospholipase D1, phosphatidylcholine-specific | Affinity Capture-Western Phenotypic Suppression Reconstituted Complex | BioGRID | 10764771 |
| PLD2 | - | phospholipase D2 | - | HPRD,BioGRID | 10764771 |
| PTK2 | FADK | FAK | FAK1 | pp125FAK | PTK2 protein tyrosine kinase 2 | Affinity Capture-Western | BioGRID | 12558988 |
| SH3GL2 | CNSA2 | EEN-B1 | FLJ20276 | FLJ25015 | SH3D2A | SH3P4 | SH3-domain GRB2-like 2 | Reconstituted Complex | BioGRID | 9341169 |12456676 |
| SH3GL2 | CNSA2 | EEN-B1 | FLJ20276 | FLJ25015 | SH3D2A | SH3P4 | SH3-domain GRB2-like 2 | - | HPRD | 9315708 |
| SH3GLB1 | Bif-1 | CGI-61 | KIAA0491 | dJ612B15.2 | SH3-domain GRB2-like endophilin B1 | - | HPRD,BioGRID | 12456676 |
| SYN1 | SYN1a | SYN1b | SYNI | synapsin I | - | HPRD,BioGRID | 10899172 |
| SYNJ1 | INPP5G | synaptojanin 1 | - | HPRD | 10542231 |14704270 |
| SYNJ1 | INPP5G | synaptojanin 1 | Affinity Capture-Western Reconstituted Complex | BioGRID | 9341169 |
| VTN | V75 | VN | VNT | vitronectin | - | HPRD,BioGRID | 12565847 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG FC GAMMA R MEDIATED PHAGOCYTOSIS | 97 | 71 | All SZGR 2.0 genes in this pathway |
| BIOCARTA NDKDYNAMIN PATHWAY | 21 | 15 | All SZGR 2.0 genes in this pathway |
| PID ERBB1 INTERNALIZATION PATHWAY | 41 | 35 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 TTD DN | 84 | 63 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| MOHANKUMAR TLX1 TARGETS DN | 193 | 112 | All SZGR 2.0 genes in this pathway |
| SCHLESINGER H3K27ME3 IN NORMAL AND METHYLATED IN CANCER | 28 | 21 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 12HR DN | 57 | 45 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 48HR DN | 428 | 306 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| MOREAUX MULTIPLE MYELOMA BY TACI UP | 412 | 249 | All SZGR 2.0 genes in this pathway |
| WEIGEL OXIDATIVE STRESS RESPONSE | 35 | 28 | All SZGR 2.0 genes in this pathway |
| ZHENG BOUND BY FOXP3 | 491 | 310 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| ZHENG FOXP3 TARGETS IN THYMUS DN | 12 | 9 | All SZGR 2.0 genes in this pathway |
| TAVAZOIE METASTASIS | 108 | 68 | All SZGR 2.0 genes in this pathway |
| CROONQUIST NRAS VS STROMAL STIMULATION UP | 41 | 26 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS CALB1 CORR UP | 548 | 370 | All SZGR 2.0 genes in this pathway |
| HOELZEL NF1 TARGETS DN | 115 | 73 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-1/206 | 1073 | 1079 | 1A | hsa-miR-1 | UGGAAUGUAAAGAAGUAUGUA |
| hsa-miR-206SZ | UGGAAUGUAAGGAAGUGUGUGG | ||||
| hsa-miR-613 | AGGAAUGUUCCUUCUUUGCC | ||||
| miR-153 | 977 | 983 | 1A | hsa-miR-153 | UUGCAUAGUCACAAAAGUGA |
| miR-26 | 259 | 265 | m8 | hsa-miR-26abrain | UUCAAGUAAUCCAGGAUAGGC |
| hsa-miR-26bSZ | UUCAAGUAAUUCAGGAUAGGUU | ||||
| miR-28 | 947 | 953 | m8 | hsa-miR-28brain | AAGGAGCUCACAGUCUAUUGAG |
| miR-330 | 231 | 237 | m8 | hsa-miR-330brain | GCAAAGCACACGGCCUGCAGAGA |
| miR-448 | 977 | 983 | 1A | hsa-miR-448 | UUGCAUAUGUAGGAUGUCCCAU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.