Gene Page: GNA11
Summary ?
| GeneID | 2767 |
| Symbol | GNA11 |
| Synonyms | FBH|FBH2|FHH2|GNA-11|HHC2|HYPOC2 |
| Description | G protein subunit alpha 11 |
| Reference | MIM:139313|HGNC:HGNC:4379|Ensembl:ENSG00000088256|HPRD:00759|Vega:OTTHUMG00000180631 |
| Gene type | protein-coding |
| Map location | 19p13.3 |
| Pascal p-value | 0.702 |
| Sherlock p-value | 0.388 |
| Fetal beta | 0.708 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
| Support | G-PROTEIN RELAY METABOTROPIC GLUTAMATE RECEPTOR |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.6384 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg02261780 | 19 | 3097728 | GNA11 | 1.98E-4 | 0.323 | 0.035 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs17542473 | chr4 | 111182451 | GNA11 | 2767 | 0.14 | trans |
Section II. Transcriptome annotation
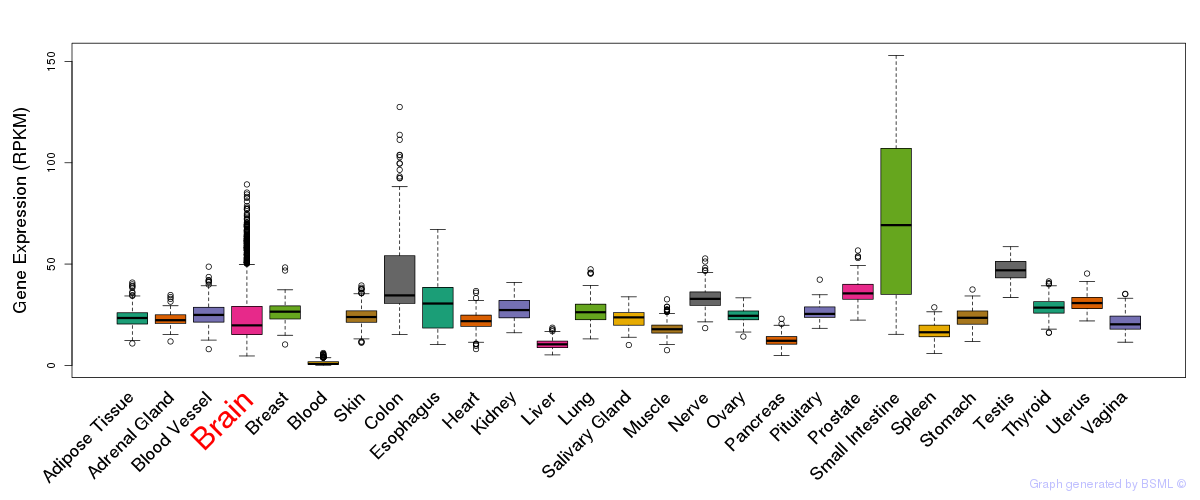
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MAMDC4 | 0.91 | 0.85 |
| PLA2G4B | 0.91 | 0.89 |
| CCDC57 | 0.91 | 0.88 |
| JMJD7-PLA2G4B | 0.90 | 0.82 |
| NEIL1 | 0.89 | 0.84 |
| CSAD | 0.89 | 0.86 |
| TGM1 | 0.89 | 0.81 |
| CCNL2 | 0.88 | 0.81 |
| AC135050.2 | 0.88 | 0.81 |
| AC011511.1 | 0.88 | 0.82 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ITM2A | -0.45 | -0.50 |
| HLA-C | -0.45 | -0.40 |
| ABCG2 | -0.43 | -0.41 |
| B2M | -0.42 | -0.42 |
| TM4SF1 | -0.42 | -0.42 |
| ITM2B | -0.42 | -0.40 |
| AF347015.31 | -0.42 | -0.44 |
| TM4SF18 | -0.42 | -0.42 |
| SRGN | -0.41 | -0.43 |
| FAM162A | -0.41 | -0.38 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0003774 | motor activity | IEA | - | |
| GO:0004871 | signal transducer activity | IEA | - | |
| GO:0003924 | GTPase activity | TAS | 8530500 | |
| GO:0005525 | GTP binding | IEA | - | |
| GO:0019001 | guanyl nucleotide binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0060158 | dopamine receptor, phospholipase C activating pathway | IEA | dopamine (GO term level: 10) | - |
| GO:0001501 | skeletal system development | IEA | - | |
| GO:0001508 | regulation of action potential | IEA | - | |
| GO:0007186 | G-protein coupled receptor protein signaling pathway | IEA | - | |
| GO:0006471 | protein amino acid ADP-ribosylation | IEA | - | |
| GO:0007165 | signal transduction | TAS | 8530500 | |
| GO:0048066 | pigmentation during development | IEA | - | |
| GO:0007507 | heart development | IEA | - | |
| GO:0006941 | striated muscle contraction | IEA | - | |
| GO:0045634 | regulation of melanocyte differentiation | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005863 | striated muscle thick filament | IEA | - | |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0005737 | cytoplasm | TAS | 9175863 | |
| GO:0005886 | plasma membrane | TAS | 9175863 | |
| GO:0016459 | myosin complex | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ADRA1B | ADRA1 | ALPHA1BAR | adrenergic, alpha-1B-, receptor | - | HPRD | 9481484 |
| ADRB2 | ADRB2R | ADRBR | B2AR | BAR | BETA2AR | adrenergic, beta-2-, receptor, surface | - | HPRD | 7797501 |
| CCR2 | CC-CKR-2 | CCR2A | CCR2B | CD192 | CKR2 | CKR2A | CKR2B | CMKBR2 | MCP-1-R | chemokine (C-C motif) receptor 2 | Affinity Capture-Western | BioGRID | 11350939 |
| CHRM2 | FLJ43243 | HM2 | MGC120006 | MGC120007 | cholinergic receptor, muscarinic 2 | - | HPRD | 7797501 |
| EDNRA | ETA | ETRA | endothelin receptor type A | - | HPRD,BioGRID | 10789830 |
| EDNRB | ABCDS | ETB | ETBR | ETRB | HSCR | HSCR2 | endothelin receptor type B | - | HPRD,BioGRID | 10789830 |
| HTR2A | 5-HT2A | HTR2 | 5-hydroxytryptamine (serotonin) receptor 2A | - | HPRD | 11916537 |
| MTNR1A | MEL-1A-R | MT1 | melatonin receptor 1A | - | HPRD,BioGRID | 10598579 |
| PTGIR | IP | MGC102830 | PRIPR | prostaglandin I2 (prostacyclin) receptor (IP) | - | HPRD | 10446129 |
| RGS3 | C2PA | FLJ20370 | FLJ31516 | FLJ90496 | PDZ-RGS3 | RGP3 | regulator of G-protein signaling 3 | - | HPRD,BioGRID | 9858594 |
| TBXA2R | TXA2-R | thromboxane A2 receptor | - | HPRD | 9152406 |
| TRPC3 | TRP3 | transient receptor potential cation channel, subfamily C, member 3 | - | HPRD | 15623527 |
| TRPC3 | TRP3 | transient receptor potential cation channel, subfamily C, member 3 | - | HPRD | 11524429 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CALCIUM SIGNALING PATHWAY | 178 | 134 | All SZGR 2.0 genes in this pathway |
| KEGG VASCULAR SMOOTH MUSCLE CONTRACTION | 115 | 81 | All SZGR 2.0 genes in this pathway |
| KEGG GAP JUNCTION | 90 | 68 | All SZGR 2.0 genes in this pathway |
| KEGG LONG TERM DEPRESSION | 70 | 53 | All SZGR 2.0 genes in this pathway |
| KEGG GNRH SIGNALING PATHWAY | 101 | 77 | All SZGR 2.0 genes in this pathway |
| BIOCARTA BIOPEPTIDES PATHWAY | 45 | 35 | All SZGR 2.0 genes in this pathway |
| ST ADRENERGIC | 36 | 29 | All SZGR 2.0 genes in this pathway |
| PID ENDOTHELIN PATHWAY | 63 | 52 | All SZGR 2.0 genes in this pathway |
| PID LYSOPHOSPHOLIPID PATHWAY | 66 | 53 | All SZGR 2.0 genes in this pathway |
| PID ER NONGENOMIC PATHWAY | 41 | 35 | All SZGR 2.0 genes in this pathway |
| PID S1P S1P3 PATHWAY | 29 | 24 | All SZGR 2.0 genes in this pathway |
| PID ARF6 PATHWAY | 35 | 27 | All SZGR 2.0 genes in this pathway |
| PID TXA2PATHWAY | 57 | 43 | All SZGR 2.0 genes in this pathway |
| PID THROMBIN PAR4 PATHWAY | 15 | 11 | All SZGR 2.0 genes in this pathway |
| PID S1P META PATHWAY | 21 | 14 | All SZGR 2.0 genes in this pathway |
| PID THROMBIN PAR1 PATHWAY | 43 | 32 | All SZGR 2.0 genes in this pathway |
| PID S1P S1P2 PATHWAY | 24 | 19 | All SZGR 2.0 genes in this pathway |
| REACTOME GASTRIN CREB SIGNALLING PATHWAY VIA PKC AND MAPK | 205 | 136 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY GPCR | 920 | 449 | All SZGR 2.0 genes in this pathway |
| REACTOME INTEGRATION OF ENERGY METABOLISM | 120 | 84 | All SZGR 2.0 genes in this pathway |
| REACTOME G ALPHA Q SIGNALLING EVENTS | 184 | 116 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF INSULIN SECRETION | 93 | 65 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | 11 | 9 | All SZGR 2.0 genes in this pathway |
| REACTOME ADP SIGNALLING THROUGH P2RY1 | 25 | 16 | All SZGR 2.0 genes in this pathway |
| REACTOME GPCR DOWNSTREAM SIGNALING | 805 | 368 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNAL AMPLIFICATION | 31 | 19 | All SZGR 2.0 genes in this pathway |
| REACTOME THROMBOXANE SIGNALLING THROUGH TP RECEPTOR | 23 | 14 | All SZGR 2.0 genes in this pathway |
| REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | 32 | 20 | All SZGR 2.0 genes in this pathway |
| REACTOME HEMOSTASIS | 466 | 331 | All SZGR 2.0 genes in this pathway |
| REACTOME PLATELET ACTIVATION SIGNALING AND AGGREGATION | 208 | 138 | All SZGR 2.0 genes in this pathway |
| GAZDA DIAMOND BLACKFAN ANEMIA ERYTHROID DN | 493 | 298 | All SZGR 2.0 genes in this pathway |
| DEURIG T CELL PROLYMPHOCYTIC LEUKEMIA UP | 368 | 234 | All SZGR 2.0 genes in this pathway |
| WAMUNYOKOLI OVARIAN CANCER LMP UP | 265 | 158 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| OUELLET OVARIAN CANCER INVASIVE VS LMP DN | 11 | 8 | All SZGR 2.0 genes in this pathway |
| PEREZ TP53 TARGETS | 1174 | 695 | All SZGR 2.0 genes in this pathway |
| PEREZ TP63 TARGETS | 355 | 243 | All SZGR 2.0 genes in this pathway |
| PEREZ TP53 AND TP63 TARGETS | 205 | 145 | All SZGR 2.0 genes in this pathway |
| GROSS HYPOXIA VIA ELK3 UP | 209 | 139 | All SZGR 2.0 genes in this pathway |
| GROSS HYPOXIA VIA ELK3 AND HIF1A DN | 103 | 71 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS DN | 848 | 527 | All SZGR 2.0 genes in this pathway |
| GOTZMANN EPITHELIAL TO MESENCHYMAL TRANSITION DN | 206 | 136 | All SZGR 2.0 genes in this pathway |
| KENNY CTNNB1 TARGETS UP | 50 | 30 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS PLASMABLAST UP | 398 | 262 | All SZGR 2.0 genes in this pathway |
| SANSOM APC TARGETS DN | 366 | 238 | All SZGR 2.0 genes in this pathway |
| MOREAUX MULTIPLE MYELOMA BY TACI UP | 412 | 249 | All SZGR 2.0 genes in this pathway |
| MOREAUX B LYMPHOCYTE MATURATION BY TACI UP | 92 | 58 | All SZGR 2.0 genes in this pathway |
| ZAMORA NOS2 TARGETS DN | 96 | 71 | All SZGR 2.0 genes in this pathway |
| AMUNDSON POOR SURVIVAL AFTER GAMMA RADIATION 8G | 95 | 62 | All SZGR 2.0 genes in this pathway |
| RIZKI TUMOR INVASIVENESS 2D UP | 69 | 46 | All SZGR 2.0 genes in this pathway |
| AMBROSINI FLAVOPIRIDOL TREATMENT TP53 | 109 | 63 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER DN | 540 | 340 | All SZGR 2.0 genes in this pathway |
| GRADE COLON AND RECTAL CANCER DN | 101 | 65 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| SHEDDEN LUNG CANCER GOOD SURVIVAL A5 | 70 | 32 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE UP | 857 | 456 | All SZGR 2.0 genes in this pathway |
| LU EZH2 TARGETS UP | 295 | 155 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP B | 549 | 316 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS UP | 682 | 433 | All SZGR 2.0 genes in this pathway |
| FOSTER KDM1A TARGETS DN | 211 | 119 | All SZGR 2.0 genes in this pathway |