Gene Page: ANG
Summary ?
| GeneID | 283 |
| Symbol | ANG |
| Synonyms | ALS9|HEL168|RAA1|RNASE4|RNASE5 |
| Description | angiogenin, ribonuclease, RNase A family, 5 |
| Reference | MIM:105850|HGNC:HGNC:483|Ensembl:ENSG00000214274|HPRD:00105|Vega:OTTHUMG00000029576 |
| Gene type | protein-coding |
| Map location | 14q11.1-q11.2 |
| Sherlock p-value | 0.091 |
| Fetal beta | 0.286 |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.047 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias,schizotypal | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 3 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs10958910 | chr9 | 10134759 | ANG | 283 | 0.15 | trans |
Section II. Transcriptome annotation
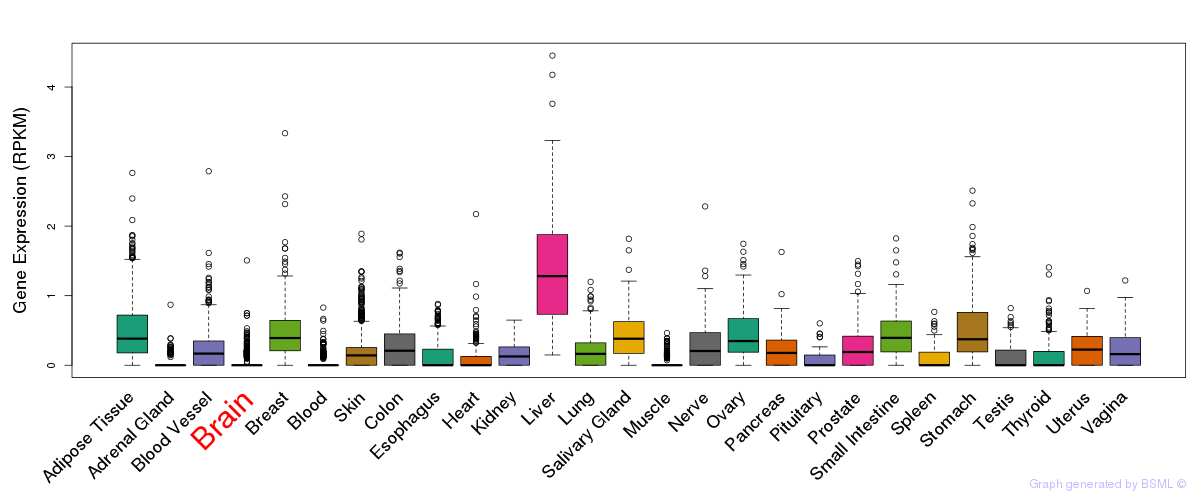
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| KCNC1 | 0.82 | 0.85 |
| SLC45A4 | 0.82 | 0.83 |
| STX1B | 0.78 | 0.82 |
| RNF157 | 0.77 | 0.82 |
| KCNC3 | 0.77 | 0.72 |
| ATP2B3 | 0.77 | 0.82 |
| KLC2 | 0.77 | 0.79 |
| CAMTA1 | 0.76 | 0.79 |
| IFFO1 | 0.76 | 0.78 |
| STXBP1 | 0.76 | 0.81 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| RHOC | -0.45 | -0.56 |
| DBI | -0.43 | -0.53 |
| GNG11 | -0.43 | -0.54 |
| C1orf54 | -0.42 | -0.56 |
| C1orf61 | -0.42 | -0.55 |
| AF347015.21 | -0.41 | -0.48 |
| RAB34 | -0.39 | -0.47 |
| MYL12A | -0.38 | -0.48 |
| VAMP5 | -0.37 | -0.46 |
| IL32 | -0.37 | -0.47 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005102 | receptor binding | IDA | Neurotransmitter (GO term level: 4) | 9122172 |
| GO:0003779 | actin binding | IDA | 7679494 |11782452 | |
| GO:0003676 | nucleic acid binding | IEA | - | |
| GO:0003677 | DNA binding | IC | 10649442 | |
| GO:0005507 | copper ion binding | IDA | 9245697 | |
| GO:0004540 | ribonuclease activity | IDA | 2424496 |2730651 | |
| GO:0004519 | endonuclease activity | IEA | - | |
| GO:0004519 | endonuclease activity | TAS | 10103013 | |
| GO:0004522 | pancreatic ribonuclease activity | IEA | - | |
| GO:0016787 | hydrolase activity | IEA | - | |
| GO:0008201 | heparin binding | IDA | 10103013 | |
| GO:0019843 | rRNA binding | TAS | 2457905 | |
| GO:0042277 | peptide binding | IDA | 11782452 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0001556 | oocyte maturation | NAS | 11438326 | |
| GO:0001525 | angiogenesis | IMP | 2479414 |17125737 | |
| GO:0001525 | angiogenesis | TAS | 16567967 | |
| GO:0001666 | response to hypoxia | IDA | 10999833 |15979542 |16490744 | |
| GO:0001666 | response to hypoxia | NAS | 15776477 | |
| GO:0001541 | ovarian follicle development | NAS | 12770725 | |
| GO:0001890 | placenta development | NAS | 11984825 | |
| GO:0001938 | positive regulation of endothelial cell proliferation | IDA | 9122172 |9707554 | |
| GO:0007202 | activation of phospholipase C activity | IMP | 2457905 | |
| GO:0007154 | cell communication | NAS | 10103013 | |
| GO:0009303 | rRNA transcription | IMP | 15735021 | |
| GO:0009725 | response to hormone stimulus | IDA | 10999833 | |
| GO:0006651 | diacylglycerol biosynthetic process | IDA | 2457905 | |
| GO:0007275 | multicellular organismal development | IEA | - | |
| GO:0042327 | positive regulation of phosphorylation | IDA | 17125737 | |
| GO:0016477 | cell migration | IMP | 17125737 | |
| GO:0030154 | cell differentiation | IEA | - | |
| GO:0042592 | homeostatic process | NAS | 15166501 | |
| GO:0030041 | actin filament polymerization | ISS | - | |
| GO:0032431 | activation of phospholipase A2 | IMP | 2646638 | |
| GO:0032148 | activation of protein kinase B activity | IMP | 17125737 | |
| GO:0017148 | negative regulation of translation | IEA | - | |
| GO:0050714 | positive regulation of protein secretion | IDA | 2646638 | |
| GO:0050714 | positive regulation of protein secretion | ISS | - | |
| GO:0048662 | negative regulation of smooth muscle cell proliferation | IDA | 10103013 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0043025 | cell soma | ISS | axon, dendrite (GO term level: 4) | - |
| GO:0030426 | growth cone | ISS | axon, dendrite (GO term level: 5) | - |
| GO:0005576 | extracellular region | IEA | - | |
| GO:0005605 | basal lamina | IDA | 15166501 | |
| GO:0005615 | extracellular space | IDA | 3663649 |16461950 |16490744 | |
| GO:0005634 | nucleus | IDA | 10649442 |15735021 | |
| GO:0005730 | nucleolus | ISS | - | |
| GO:0032311 | angiogenin-PRI complex | IPI | 3470787 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| BARRIER COLON CANCER RECURRENCE UP | 42 | 28 | All SZGR 2.0 genes in this pathway |
| ELVIDGE HYPOXIA UP | 171 | 112 | All SZGR 2.0 genes in this pathway |
| ELVIDGE HYPOXIA BY DMOG UP | 130 | 85 | All SZGR 2.0 genes in this pathway |
| ELVIDGE HIF1A TARGETS DN | 91 | 58 | All SZGR 2.0 genes in this pathway |
| ELVIDGE HIF1A AND HIF2A TARGETS DN | 104 | 72 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE EPIDERMIS DN | 508 | 354 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS G UP | 238 | 135 | All SZGR 2.0 genes in this pathway |
| RASHI RESPONSE TO IONIZING RADIATION 5 | 147 | 89 | All SZGR 2.0 genes in this pathway |
| RANKIN ANGIOGENIC TARGETS OF VHL HIF2A UP | 5 | 5 | All SZGR 2.0 genes in this pathway |
| RICKMAN METASTASIS UP | 344 | 180 | All SZGR 2.0 genes in this pathway |
| GEORGES TARGETS OF MIR192 AND MIR215 | 893 | 528 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 1 UP | 140 | 85 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS PLASMABLAST DN | 309 | 206 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| SANSOM APC TARGETS UP | 126 | 84 | All SZGR 2.0 genes in this pathway |
| HSIAO LIVER SPECIFIC GENES | 244 | 154 | All SZGR 2.0 genes in this pathway |
| MENSE HYPOXIA UP | 98 | 71 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL CULTURED VS FRESH UP | 425 | 298 | All SZGR 2.0 genes in this pathway |
| VART KSHV INFECTION ANGIOGENIC MARKERS DN | 138 | 92 | All SZGR 2.0 genes in this pathway |
| MIZUKAMI HYPOXIA UP | 12 | 12 | All SZGR 2.0 genes in this pathway |
| CHEN METABOLIC SYNDROM NETWORK | 1210 | 725 | All SZGR 2.0 genes in this pathway |
| ROME INSULIN TARGETS IN MUSCLE DN | 204 | 114 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA DN | 267 | 160 | All SZGR 2.0 genes in this pathway |
| SERVITJA LIVER HNF1A TARGETS DN | 157 | 105 | All SZGR 2.0 genes in this pathway |
| KRIEG HYPOXIA NOT VIA KDM3A | 770 | 480 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 1ST EGF PULSE ONLY | 1839 | 928 | All SZGR 2.0 genes in this pathway |