Gene Page: GRIN2D
Summary ?
| GeneID | 2906 |
| Symbol | GRIN2D |
| Synonyms | EB11|GluN2D|NMDAR2D|NR2D |
| Description | glutamate ionotropic receptor NMDA type subunit 2D |
| Reference | MIM:602717|HGNC:HGNC:4588|Ensembl:ENSG00000105464|HPRD:04095|Vega:OTTHUMG00000183304 |
| Gene type | protein-coding |
| Map location | 19q13.33 |
| Pascal p-value | 0.35 |
| Fetal beta | -0.967 |
| Support | LIGAND GATED ION SIGNALING G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS G2Cdb.human_TAP-PSD-95-CORE G2Cdb.humanPSD G2Cdb.humanPSP CompositeSet Potential synaptic genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Association | A combined odds ratio method (Sun et al. 2008), association studies | 1 | Link to SZGene |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 4 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.092 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
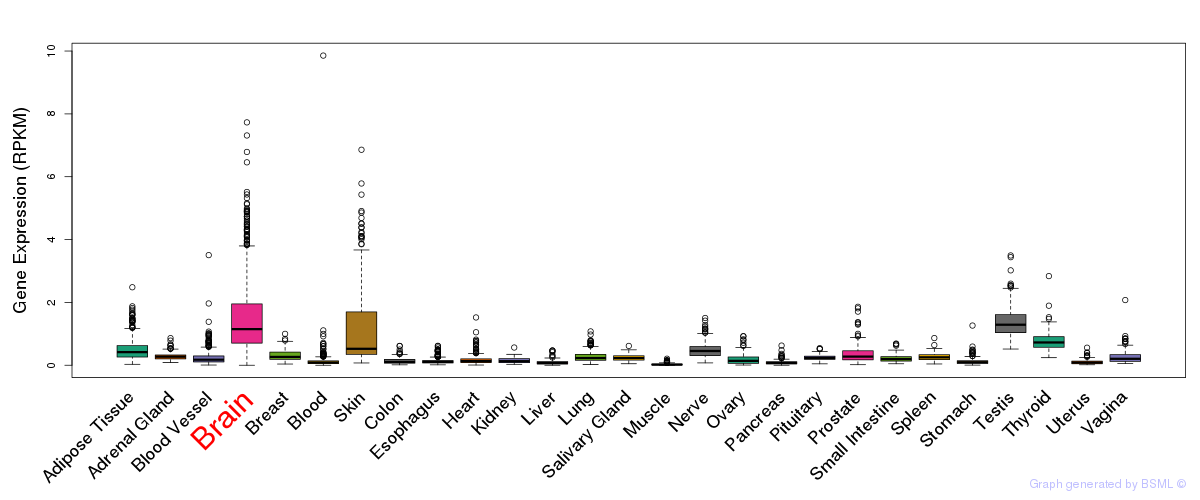
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| HSP90B1 | 0.87 | 0.85 |
| TM9SF2 | 0.86 | 0.84 |
| COPB2 | 0.86 | 0.83 |
| SRP54 | 0.86 | 0.86 |
| RAB5A | 0.86 | 0.83 |
| ATP2C1 | 0.85 | 0.84 |
| CANX | 0.85 | 0.83 |
| NIPA2 | 0.85 | 0.79 |
| CALR | 0.84 | 0.79 |
| PSMD1 | 0.83 | 0.83 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.21 | -0.69 | -0.59 |
| MT-CO2 | -0.69 | -0.66 |
| FXYD1 | -0.67 | -0.62 |
| AF347015.33 | -0.66 | -0.64 |
| AF347015.31 | -0.66 | -0.64 |
| AF347015.8 | -0.65 | -0.63 |
| AF347015.2 | -0.64 | -0.65 |
| HIGD1B | -0.64 | -0.60 |
| IFI27 | -0.63 | -0.60 |
| MT-CYB | -0.62 | -0.60 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004972 | N-methyl-D-aspartate selective glutamate receptor activity | IEA | glutamate (GO term level: 8) | - |
| GO:0005234 | extracellular-glutamate-gated ion channel activity | IEA | glutamate (GO term level: 11) | - |
| GO:0000287 | magnesium ion binding | IEA | - | |
| GO:0004872 | receptor activity | IEA | - | |
| GO:0005509 | calcium ion binding | IEA | - | |
| GO:0005216 | ion channel activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0001964 | startle response | IEA | - | |
| GO:0007165 | signal transduction | NAS | 9480759 | |
| GO:0008344 | adult locomotory behavior | IEA | - | |
| GO:0006811 | ion transport | IEA | - | |
| GO:0051930 | regulation of sensory perception of pain | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0019717 | synaptosome | IEA | Synap, Brain (GO term level: 7) | - |
| GO:0045211 | postsynaptic membrane | IEA | Synap, Neurotransmitter (GO term level: 5) | - |
| GO:0045202 | synapse | IEA | neuron, Synap, Neurotransmitter, Glial (GO term level: 2) | - |
| GO:0016021 | integral to membrane | IEA | - | |
| GO:0005886 | plasma membrane | IEA | - | |
| GO:0030054 | cell junction | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ABL1 | ABL | JTK7 | bcr/abl | c-ABL | p150 | v-abl | c-abl oncogene 1, receptor tyrosine kinase | - | HPRD | 10777567 |
| DLG4 | FLJ97752 | FLJ98574 | PSD95 | SAP90 | discs, large homolog 4 (Drosophila) | - | HPRD,BioGRID | 7569905 |
| DLGAP4 | DAP4 | KIAA0964 | MGC131862 | RP5-977B1.6 | SAPAP4 | discs, large (Drosophila) homolog-associated protein 4 | - | HPRD | 10862698 |
| DNM1 | DNM | dynamin 1 | - | HPRD | 10862698 |
| DUSP4 | HVH2 | MKP-2 | MKP2 | TYP | dual specificity phosphatase 4 | - | HPRD | 10862698 |
| GRIN1 | NMDA1 | NMDAR1 | NR1 | glutamate receptor, ionotropic, N-methyl D-aspartate 1 | - | HPRD | 12191494 |
| IL16 | FLJ16806 | FLJ42735 | FLJ44234 | HsT19289 | IL-16 | LCF | prIL-16 | interleukin 16 (lymphocyte chemoattractant factor) | - | HPRD,BioGRID | 10479680 |
| INADL | Cipp | FLJ26982 | InaD-like | PATJ | InaD-like (Drosophila) | - | HPRD | 9647694 |
| MAP2 | DKFZp686I2148 | MAP2A | MAP2B | MAP2C | microtubule-associated protein 2 | - | HPRD | 10862698 |
| NANOS1 | NOS1 | nanos homolog 1 (Drosophila) | - | HPRD | 10862698 |
| PPP2R1A | MGC786 | PR65A | protein phosphatase 2 (formerly 2A), regulatory subunit A, alpha isoform | - | HPRD | 10862698 |
| SYNGAP1 | DKFZp761G1421 | KIAA1938 | RASA1 | RASA5 | SYNGAP | synaptic Ras GTPase activating protein 1 homolog (rat) | - | HPRD | 10862698 |
| TJP1 | DKFZp686M05161 | MGC133289 | ZO-1 | tight junction protein 1 (zona occludens 1) | - | HPRD | 10862698 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CALCIUM SIGNALING PATHWAY | 178 | 134 | All SZGR 2.0 genes in this pathway |
| KEGG NEUROACTIVE LIGAND RECEPTOR INTERACTION | 272 | 195 | All SZGR 2.0 genes in this pathway |
| KEGG LONG TERM POTENTIATION | 70 | 57 | All SZGR 2.0 genes in this pathway |
| KEGG ALZHEIMERS DISEASE | 169 | 110 | All SZGR 2.0 genes in this pathway |
| KEGG AMYOTROPHIC LATERAL SCLEROSIS ALS | 53 | 43 | All SZGR 2.0 genes in this pathway |
| BIOCARTA NOS1 PATHWAY | 24 | 21 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSMISSION ACROSS CHEMICAL SYNAPSES | 186 | 155 | All SZGR 2.0 genes in this pathway |
| REACTOME NEURONAL SYSTEM | 279 | 221 | All SZGR 2.0 genes in this pathway |
| REACTOME NEUROTRANSMITTER RECEPTOR BINDING AND DOWNSTREAM TRANSMISSION IN THE POSTSYNAPTIC CELL | 137 | 110 | All SZGR 2.0 genes in this pathway |
| REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | 17 | 15 | All SZGR 2.0 genes in this pathway |
| REACTOME ACTIVATION OF NMDA RECEPTOR UPON GLUTAMATE BINDING AND POSTSYNAPTIC EVENTS | 37 | 32 | All SZGR 2.0 genes in this pathway |
| REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF RAS | 27 | 23 | All SZGR 2.0 genes in this pathway |
| REACTOME POST NMDA RECEPTOR ACTIVATION EVENTS | 33 | 28 | All SZGR 2.0 genes in this pathway |
| REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | 15 | 14 | All SZGR 2.0 genes in this pathway |
| REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | 15 | 14 | All SZGR 2.0 genes in this pathway |
| BENPORATH SUZ12 TARGETS | 1038 | 678 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| MASSARWEH TAMOXIFEN RESISTANCE UP | 578 | 341 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3K4ME2 | 491 | 319 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE LATE | 1137 | 655 | All SZGR 2.0 genes in this pathway |