Gene Page: HEXB
Summary ?
| GeneID | 3074 |
| Symbol | HEXB |
| Synonyms | ENC-1AS|HEL-248|HEL-S-111 |
| Description | hexosaminidase subunit beta |
| Reference | MIM:606873|HGNC:HGNC:4879|Ensembl:ENSG00000049860|HPRD:06043|Vega:OTTHUMG00000102057 |
| Gene type | protein-coding |
| Map location | 5q13 |
| Pascal p-value | 0.858 |
| Sherlock p-value | 0.554 |
| Fetal beta | -1.327 |
| eGene | Caudate basal ganglia Nucleus accumbens basal ganglia Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
General gene expression (GTEx)
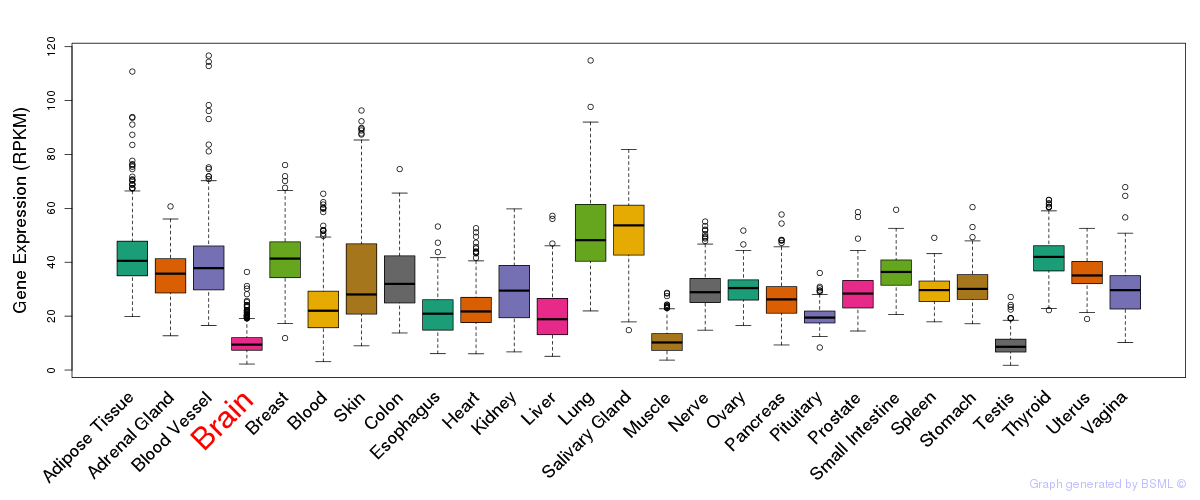
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ANKFY1 | 0.86 | 0.89 |
| ZNF592 | 0.85 | 0.87 |
| DIP2A | 0.85 | 0.90 |
| MTF1 | 0.85 | 0.89 |
| ETV3 | 0.85 | 0.86 |
| ZKSCAN1 | 0.85 | 0.91 |
| SPAG9 | 0.84 | 0.88 |
| KSR1 | 0.84 | 0.88 |
| BICD2 | 0.84 | 0.89 |
| KIAA0355 | 0.84 | 0.88 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.21 | -0.62 | -0.73 |
| AF347015.31 | -0.61 | -0.68 |
| C1orf54 | -0.60 | -0.72 |
| HIGD1B | -0.59 | -0.66 |
| MT-CO2 | -0.59 | -0.66 |
| ENHO | -0.58 | -0.65 |
| IFI27 | -0.58 | -0.62 |
| FXYD1 | -0.58 | -0.63 |
| VAMP5 | -0.57 | -0.63 |
| MT3 | -0.57 | -0.63 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004563 | beta-N-acetylhexosaminidase activity | TAS | 8663217 | |
| GO:0016798 | hydrolase activity, acting on glycosyl bonds | IEA | - | |
| GO:0042803 | protein homodimerization activity | IDA | 6230359 | |
| GO:0043169 | cation binding | IEA | - | |
| GO:0046982 | protein heterodimerization activity | IDA | 6230359 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0042552 | myelination | IEA | neuron, axon, Brain, oligodendrocyte (GO term level: 13) | - |
| GO:0001501 | skeletal system development | IEA | - | |
| GO:0008654 | phospholipid biosynthetic process | IEA | - | |
| GO:0005975 | carbohydrate metabolic process | IEA | - | |
| GO:0048477 | oogenesis | IEA | - | |
| GO:0008152 | metabolic process | IEA | - | |
| GO:0008049 | male courtship behavior | IEA | - | |
| GO:0019953 | sexual reproduction | IEA | - | |
| GO:0007626 | locomotory behavior | IEA | - | |
| GO:0007605 | sensory perception of sound | IEA | - | |
| GO:0019915 | sequestering of lipid | IEA | - | |
| GO:0009313 | oligosaccharide catabolic process | IEA | - | |
| GO:0007040 | lysosome organization | IEA | - | |
| GO:0006689 | ganglioside catabolic process | IEA | - | |
| GO:0006687 | glycosphingolipid metabolic process | IEA | - | |
| GO:0007341 | penetration of zona pellucida | IEA | - | |
| GO:0006874 | cellular calcium ion homeostasis | IEA | - | |
| GO:0030203 | glycosaminoglycan metabolic process | IEA | - | |
| GO:0050885 | neuromuscular process controlling balance | IEA | - | |
| GO:0031323 | regulation of cellular metabolic process | IEA | - | |
| GO:0044267 | cellular protein metabolic process | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0001669 | acrosome | IEA | - | |
| GO:0016020 | membrane | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG OTHER GLYCAN DEGRADATION | 16 | 12 | All SZGR 2.0 genes in this pathway |
| KEGG AMINO SUGAR AND NUCLEOTIDE SUGAR METABOLISM | 44 | 30 | All SZGR 2.0 genes in this pathway |
| KEGG GLYCOSAMINOGLYCAN DEGRADATION | 21 | 14 | All SZGR 2.0 genes in this pathway |
| KEGG GLYCOSPHINGOLIPID BIOSYNTHESIS GLOBO SERIES | 14 | 12 | All SZGR 2.0 genes in this pathway |
| KEGG GLYCOSPHINGOLIPID BIOSYNTHESIS GANGLIO SERIES | 15 | 9 | All SZGR 2.0 genes in this pathway |
| KEGG LYSOSOME | 121 | 83 | All SZGR 2.0 genes in this pathway |
| REACTOME GLYCOSPHINGOLIPID METABOLISM | 38 | 22 | All SZGR 2.0 genes in this pathway |
| REACTOME PHOSPHOLIPID METABOLISM | 198 | 112 | All SZGR 2.0 genes in this pathway |
| REACTOME CS DS DEGRADATION | 12 | 9 | All SZGR 2.0 genes in this pathway |
| REACTOME HYALURONAN UPTAKE AND DEGRADATION | 10 | 7 | All SZGR 2.0 genes in this pathway |
| REACTOME HYALURONAN METABOLISM | 14 | 7 | All SZGR 2.0 genes in this pathway |
| REACTOME CHONDROITIN SULFATE DERMATAN SULFATE METABOLISM | 49 | 33 | All SZGR 2.0 genes in this pathway |
| REACTOME KERATAN SULFATE KERATIN METABOLISM | 30 | 21 | All SZGR 2.0 genes in this pathway |
| REACTOME KERATAN SULFATE DEGRADATION | 11 | 7 | All SZGR 2.0 genes in this pathway |
| REACTOME GLYCOSAMINOGLYCAN METABOLISM | 111 | 69 | All SZGR 2.0 genes in this pathway |
| REACTOME SPHINGOLIPID METABOLISM | 69 | 40 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF LIPIDS AND LIPOPROTEINS | 478 | 302 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF CARBOHYDRATES | 247 | 154 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 1 UP | 380 | 236 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 2 UP | 418 | 263 | All SZGR 2.0 genes in this pathway |
| TIEN INTESTINE PROBIOTICS 6HR DN | 167 | 100 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN UP | 1142 | 669 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN UP | 612 | 367 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE DN | 712 | 443 | All SZGR 2.0 genes in this pathway |
| LI WILMS TUMOR VS FETAL KIDNEY 2 UP | 30 | 24 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS B LYMPHOCYTE UP | 78 | 51 | All SZGR 2.0 genes in this pathway |
| ROSS AML WITH MLL FUSIONS | 78 | 45 | All SZGR 2.0 genes in this pathway |
| BROWN MYELOID CELL DEVELOPMENT UP | 165 | 100 | All SZGR 2.0 genes in this pathway |
| APPEL IMATINIB RESPONSE | 33 | 22 | All SZGR 2.0 genes in this pathway |
| REN ALVEOLAR RHABDOMYOSARCOMA DN | 408 | 274 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| DAZARD RESPONSE TO UV NHEK UP | 244 | 151 | All SZGR 2.0 genes in this pathway |
| HENDRICKS SMARCA4 TARGETS DN | 53 | 34 | All SZGR 2.0 genes in this pathway |
| LEE AGING CEREBELLUM UP | 84 | 58 | All SZGR 2.0 genes in this pathway |
| RODWELL AGING KIDNEY NO BLOOD UP | 222 | 139 | All SZGR 2.0 genes in this pathway |
| WELCSH BRCA1 TARGETS UP | 198 | 132 | All SZGR 2.0 genes in this pathway |
| JIANG AGING HYPOTHALAMUS UP | 47 | 31 | All SZGR 2.0 genes in this pathway |
| MONNIER POSTRADIATION TUMOR ESCAPE DN | 373 | 196 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE BY GAMMA AND UV RADIATION | 88 | 65 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE DN | 195 | 135 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO CSF2RB AND IL4 DN | 315 | 201 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF UP | 418 | 282 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF VS CSF2RB AND IL4 UP | 408 | 276 | All SZGR 2.0 genes in this pathway |
| ROSS LEUKEMIA WITH MLL FUSIONS | 78 | 49 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS 60HR UP | 293 | 203 | All SZGR 2.0 genes in this pathway |
| COLINA TARGETS OF 4EBP1 AND 4EBP2 | 356 | 214 | All SZGR 2.0 genes in this pathway |
| CHEN METABOLIC SYNDROM NETWORK | 1210 | 725 | All SZGR 2.0 genes in this pathway |
| ZHAN LATE DIFFERENTIATION GENES UP | 33 | 24 | All SZGR 2.0 genes in this pathway |
| VERHAAK GLIOBLASTOMA MESENCHYMAL | 216 | 130 | All SZGR 2.0 genes in this pathway |
| HIRSCH CELLULAR TRANSFORMATION SIGNATURE UP | 242 | 159 | All SZGR 2.0 genes in this pathway |
| WANG RESPONSE TO GSK3 INHIBITOR SB216763 UP | 397 | 206 | All SZGR 2.0 genes in this pathway |
| RAO BOUND BY SALL4 | 227 | 149 | All SZGR 2.0 genes in this pathway |