Gene Page: IGF2
Summary ?
| GeneID | 3481 |
| Symbol | IGF2 |
| Synonyms | C11orf43|GRDF|IGF-II|PP9974 |
| Description | insulin like growth factor 2 |
| Reference | MIM:147470|HGNC:HGNC:5466|Ensembl:ENSG00000167244|HPRD:00939|HPRD:17410|Vega:OTTHUMG00000009395 |
| Gene type | protein-coding |
| Map location | 11p15.5 |
| Pascal p-value | 0.881 |
| Fetal beta | -0.11 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
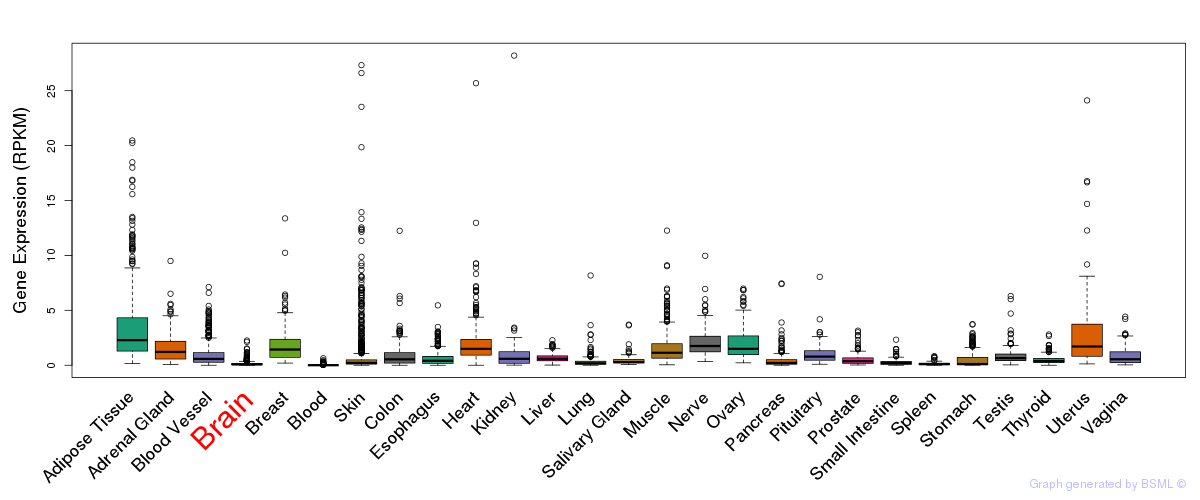
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005179 | hormone activity | IEA | - | |
| GO:0005159 | insulin-like growth factor receptor binding | TAS | 1845984 | |
| GO:0005158 | insulin receptor binding | IPI | 12138094 | |
| GO:0005515 | protein binding | IEA | - | |
| GO:0008083 | growth factor activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0001501 | skeletal system development | TAS | 8298652 | |
| GO:0006349 | genetic imprinting | TAS | 8968759 | |
| GO:0009887 | organ morphogenesis | IEA | - | |
| GO:0008283 | cell proliferation | IEA | - | |
| GO:0008286 | insulin receptor signaling pathway | TAS | 1845984 | |
| GO:0007275 | multicellular organismal development | TAS | 6382022 | |
| GO:0032583 | regulation of gene-specific transcription | NAS | 12881524 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005576 | extracellular region | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| GPC3 | DGSX | OCI-5 | SDYS | SGB | SGBS | SGBS1 | glypican 3 | - | HPRD | 8589713 |
| IDE | FLJ35968 | INSULYSIN | insulin-degrading enzyme | - | HPRD,BioGRID | 1733942 |
| IGF1R | CD221 | IGFIR | JTK13 | MGC142170 | MGC142172 | MGC18216 | insulin-like growth factor 1 receptor | - | HPRD,BioGRID | 9972281 |
| IGFBP1 | AFBP | IBP1 | IGF-BP25 | PP12 | hIGFBP-1 | insulin-like growth factor binding protein 1 | IGF-II interacts with IGFBP-1. | BIND | 10407151 |
| IGFBP1 | AFBP | IBP1 | IGF-BP25 | PP12 | hIGFBP-1 | insulin-like growth factor binding protein 1 | - | HPRD,BioGRID | 10810289 |
| IGFBP2 | IBP2 | IGF-BP53 | insulin-like growth factor binding protein 2, 36kDa | IGF-II interacts with IGFBP-2. | BIND | 10407151 |
| IGFBP2 | IBP2 | IGF-BP53 | insulin-like growth factor binding protein 2, 36kDa | - | HPRD,BioGRID | 1714916 |2465304 |8781553 |
| IGFBP3 | BP-53 | IBP3 | insulin-like growth factor binding protein 3 | Reconstituted Complex | BioGRID | 11600567 |11749962 |
| IGFBP3 | BP-53 | IBP3 | insulin-like growth factor binding protein 3 | IGF-II interacts with IGFBP-3. | BIND | 10407151 |
| IGFBP4 | BP-4 | HT29-IGFBP | IBP4 | IGFBP-4 | insulin-like growth factor binding protein 4 | IGF-II interacts with IGFBP-4. | BIND | 10407151 |
| IGFBP4 | BP-4 | HT29-IGFBP | IBP4 | IGFBP-4 | insulin-like growth factor binding protein 4 | - | HPRD,BioGRID | 9722589 |
| IGFBP5 | IBP5 | insulin-like growth factor binding protein 5 | IGF-II interacts with IGFBP-5. | BIND | 10407151 |
| IGFBP5 | IBP5 | insulin-like growth factor binding protein 5 | - | HPRD,BioGRID | 9497324 |
| IGFBP6 | IBP6 | insulin-like growth factor binding protein 6 | IGF-II interacts with IGFBP-6. | BIND | 10407151 |
| IGFBP6 | IBP6 | insulin-like growth factor binding protein 6 | - | HPRD,BioGRID | 7683646 |
| IGFBP7 | FSTL2 | IGFBP-7 | IGFBP-7v | IGFBPRP1 | MAC25 | PSF | insulin-like growth factor binding protein 7 | - | HPRD,BioGRID | 8939990 |
| INSR | CD220 | HHF5 | insulin receptor | - | HPRD,BioGRID | 9722981 |
| NOV | CCN3 | IGFBP9 | nephroblastoma overexpressed gene | - | HPRD,BioGRID | 10084601 |
| TF | DKFZp781D0156 | PRO1557 | PRO2086 | transferrin | Reconstituted Complex | BioGRID | 11749962 |
| VTN | V75 | VN | VNT | vitronectin | - | HPRD,BioGRID | 10342887 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PID AJDISS 2PATHWAY | 48 | 38 | All SZGR 2.0 genes in this pathway |
| REACTOME DIABETES PATHWAYS | 133 | 91 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | 16 | 12 | All SZGR 2.0 genes in this pathway |
| DELYS THYROID CANCER DN | 232 | 154 | All SZGR 2.0 genes in this pathway |
| CHIARADONNA NEOPLASTIC TRANSFORMATION CDC25 UP | 120 | 73 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA REVERSIBLY DN | 29 | 21 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA UP | 536 | 340 | All SZGR 2.0 genes in this pathway |
| MISSIAGLIA REGULATED BY METHYLATION UP | 126 | 78 | All SZGR 2.0 genes in this pathway |
| OLSSON E2F3 TARGETS DN | 49 | 33 | All SZGR 2.0 genes in this pathway |
| BREUHAHN GROWTH FACTOR SIGNALING IN LIVER CANCER | 22 | 19 | All SZGR 2.0 genes in this pathway |
| LI WILMS TUMOR VS FETAL KIDNEY 2 UP | 30 | 24 | All SZGR 2.0 genes in this pathway |
| LIEN BREAST CARCINOMA METAPLASTIC | 35 | 25 | All SZGR 2.0 genes in this pathway |
| THEODOROU MAMMARY TUMORIGENESIS | 31 | 24 | All SZGR 2.0 genes in this pathway |
| KHETCHOUMIAN TRIM24 TARGETS UP | 47 | 38 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS IMPRINTED AND X LINKED | 17 | 8 | All SZGR 2.0 genes in this pathway |
| GOUYER TATI TARGETS DN | 17 | 12 | All SZGR 2.0 genes in this pathway |
| KOYAMA SEMA3B TARGETS DN | 411 | 249 | All SZGR 2.0 genes in this pathway |
| BENPORATH SUZ12 TARGETS | 1038 | 678 | All SZGR 2.0 genes in this pathway |
| BENPORATH EED TARGETS | 1062 | 725 | All SZGR 2.0 genes in this pathway |
| CHEN NEUROBLASTOMA COPY NUMBER GAINS | 50 | 35 | All SZGR 2.0 genes in this pathway |
| SHEPARD CRUSH AND BURN MUTANT UP | 197 | 110 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER MYC TGFA UP | 61 | 40 | All SZGR 2.0 genes in this pathway |
| KENNY CTNNB1 TARGETS DN | 52 | 34 | All SZGR 2.0 genes in this pathway |
| SHEPARD BMYB MORPHOLINO UP | 205 | 126 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER MYC DN | 63 | 40 | All SZGR 2.0 genes in this pathway |
| DORSAM HOXA9 TARGETS DN | 32 | 22 | All SZGR 2.0 genes in this pathway |
| GERY CEBP TARGETS | 126 | 90 | All SZGR 2.0 genes in this pathway |
| IYENGAR RESPONSE TO ADIPOCYTE FACTORS | 10 | 8 | All SZGR 2.0 genes in this pathway |
| NATSUME RESPONSE TO INTERFERON BETA UP | 71 | 49 | All SZGR 2.0 genes in this pathway |
| HARRIS HYPOXIA | 81 | 64 | All SZGR 2.0 genes in this pathway |
| INGRAM SHH TARGETS | 7 | 5 | All SZGR 2.0 genes in this pathway |
| SUZUKI RESPONSE TO TSA AND DECITABINE 1B | 23 | 14 | All SZGR 2.0 genes in this pathway |
| SATO SILENCED BY METHYLATION IN PANCREATIC CANCER 2 | 50 | 34 | All SZGR 2.0 genes in this pathway |
| DELASERNA MYOD TARGETS UP | 89 | 51 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS 5 | 126 | 78 | All SZGR 2.0 genes in this pathway |
| YAMAZAKI TCEB3 TARGETS DN | 215 | 132 | All SZGR 2.0 genes in this pathway |
| ZHU CMV 24 HR DN | 91 | 64 | All SZGR 2.0 genes in this pathway |
| ZHU CMV ALL DN | 128 | 93 | All SZGR 2.0 genes in this pathway |
| KUNINGER IGF1 VS PDGFB TARGETS UP | 82 | 51 | All SZGR 2.0 genes in this pathway |
| JACKSON DNMT1 TARGETS DN | 25 | 21 | All SZGR 2.0 genes in this pathway |
| SEMENZA HIF1 TARGETS | 36 | 32 | All SZGR 2.0 genes in this pathway |
| YIH RESPONSE TO ARSENITE C3 | 36 | 24 | All SZGR 2.0 genes in this pathway |
| JOSEPH RESPONSE TO SODIUM BUTYRATE UP | 31 | 21 | All SZGR 2.0 genes in this pathway |
| SANSOM APC MYC TARGETS | 217 | 138 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS UP | 673 | 430 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS UP | 602 | 364 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS UP | 601 | 369 | All SZGR 2.0 genes in this pathway |
| RIZKI TUMOR INVASIVENESS 2D UP | 69 | 46 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER DN | 540 | 340 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER TUMOR VS NORMAL ADJACENT TISSUE DN | 274 | 165 | All SZGR 2.0 genes in this pathway |
| WINTER HYPOXIA METAGENE | 242 | 168 | All SZGR 2.0 genes in this pathway |
| JIANG TIP30 TARGETS DN | 24 | 14 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K27ME3 | 269 | 159 | All SZGR 2.0 genes in this pathway |
| CHIANG LIVER CANCER SUBCLASS INTERFERON DN | 52 | 30 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3K4ME2 AND H3K27ME3 | 349 | 234 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN IPS WITH HCP H3K27ME3 | 102 | 76 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER SUBCLASS S2 | 115 | 74 | All SZGR 2.0 genes in this pathway |
| KYNG WERNER SYNDROM AND NORMAL AGING DN | 225 | 124 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN NPC HCP WITH H3K27ME3 | 341 | 243 | All SZGR 2.0 genes in this pathway |
| NAKAMURA ADIPOGENESIS LATE UP | 104 | 67 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS UP | 504 | 321 | All SZGR 2.0 genes in this pathway |
| JUBAN TARGETS OF SPI1 AND FLI1 DN | 92 | 60 | All SZGR 2.0 genes in this pathway |
| PLASARI TGFB1 SIGNALING VIA NFIC 1HR DN | 106 | 77 | All SZGR 2.0 genes in this pathway |
| BRIDEAU IMPRINTED GENES | 63 | 47 | All SZGR 2.0 genes in this pathway |
| NABA SECRETED FACTORS | 344 | 197 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME ASSOCIATED | 753 | 411 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME | 1028 | 559 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-141/200a | 3466 | 3473 | 1A,m8 | hsa-miR-141 | UAACACUGUCUGGUAAAGAUGG |
| hsa-miR-200a | UAACACUGUCUGGUAACGAUGU | ||||
| miR-324-3p | 2207 | 2213 | m8 | hsa-miR-324-3p | CCACUGCCCCAGGUGCUGCUGG |
| miR-544 | 2297 | 2303 | m8 | hsa-miR-544 | AUUCUGCAUUUUUAGCAAGU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.