Gene Page: IKBKB
Summary ?
| GeneID | 3551 |
| Symbol | IKBKB |
| Synonyms | IKK-beta|IKK2|IKKB|IMD15|NFKBIKB |
| Description | inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase beta |
| Reference | MIM:603258|HGNC:HGNC:5960|Ensembl:ENSG00000104365|HPRD:04462|Vega:OTTHUMG00000164092 |
| Gene type | protein-coding |
| Map location | 8p11.2 |
| Pascal p-value | 0.301 |
| Sherlock p-value | 0.506 |
| Fetal beta | 0.683 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.3858 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg11636714 | 8 | 42128950 | IKBKB | 1.087E-4 | -0.213 | 0.028 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs16829545 | chr2 | 151977407 | IKBKB | 3551 | 0 | trans | ||
| rs7584986 | chr2 | 184111432 | IKBKB | 3551 | 0.05 | trans | ||
| rs3118341 | chr9 | 25185518 | IKBKB | 3551 | 0.17 | trans | ||
| rs16955618 | chr15 | 29937543 | IKBKB | 3551 | 8.597E-6 | trans | ||
| rs16993189 | chrX | 144666166 | IKBKB | 3551 | 0.05 | trans |
Section II. Transcriptome annotation
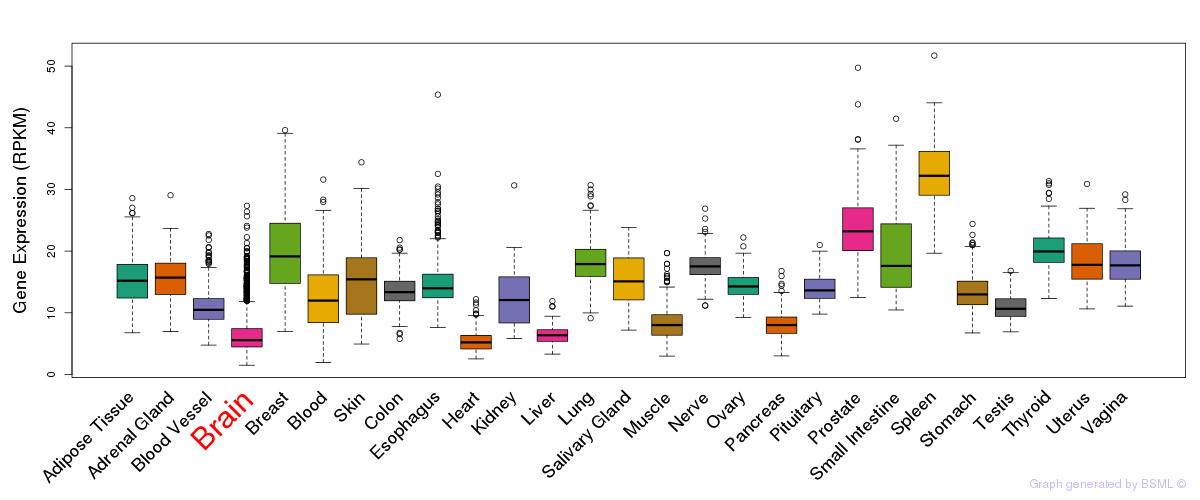
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| RGS1 | 0.72 | 0.61 |
| GPR183 | 0.70 | 0.52 |
| IFIT1 | 0.65 | 0.36 |
| MT1M | 0.64 | 0.31 |
| HIST1H1C | 0.64 | 0.28 |
| SCRG1 | 0.62 | 0.33 |
| MT1X | 0.61 | 0.32 |
| MT1F | 0.61 | 0.30 |
| MCEE | 0.61 | 0.26 |
| SAT1 | 0.60 | 0.38 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| TIAF1 | -0.39 | -0.38 |
| SURF6 | -0.38 | -0.41 |
| TNKS1BP1 | -0.37 | -0.39 |
| ZGPAT | -0.37 | -0.43 |
| MYO18A | -0.37 | -0.35 |
| KIAA0649 | -0.36 | -0.40 |
| SAFB2 | -0.36 | -0.41 |
| CENPB | -0.36 | -0.39 |
| ANKRD11 | -0.35 | -0.33 |
| SLC19A1 | -0.35 | -0.37 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0005524 | ATP binding | NAS | - | |
| GO:0004674 | protein serine/threonine kinase activity | IEA | - | |
| GO:0004713 | protein tyrosine kinase activity | IEA | - | |
| GO:0016740 | transferase activity | IEA | - | |
| GO:0008384 | IkappaB kinase activity | IEA | - | |
| GO:0016563 | transcription activator activity | NAS | 9346484 |9346485 |9813230 | |
| GO:0042802 | identical protein binding | IPI | 16319058 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0001782 | B cell homeostasis | IEA | - | |
| GO:0006464 | protein modification process | IEA | - | |
| GO:0006468 | protein amino acid phosphorylation | IEA | - | |
| GO:0006468 | protein amino acid phosphorylation | NAS | 9346484 | |
| GO:0051092 | positive regulation of NF-kappaB transcription factor activity | IDA | 15790681 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005829 | cytosol | EXP | 9744859 |10723127 | |
| GO:0005737 | cytoplasm | IDA | 18029348 | |
| GO:0005737 | cytoplasm | NAS | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| BIRC3 | AIP1 | API2 | CIAP2 | HAIP1 | HIAP1 | MALT2 | MIHC | RNF49 | baculoviral IAP repeat-containing 3 | The cIAP-2 promoter interacts with IKK-beta. | BIND | 15494311 |
| BTRC | BETA-TRCP | FBW1A | FBXW1 | FBXW1A | FWD1 | MGC4643 | bTrCP | bTrCP1 | betaTrCP | beta-transducin repeat containing | Affinity Capture-Western | BioGRID | 11158290 |
| CASP8 | ALPS2B | CAP4 | FLICE | FLJ17672 | MACH | MCH5 | MGC78473 | caspase 8, apoptosis-related cysteine peptidase | - | HPRD,BioGRID | 11002417 |
| CDC37 | P50CDC37 | cell division cycle 37 homolog (S. cerevisiae) | Co-purification Reconstituted Complex | BioGRID | 11864612 |
| CDC37 | P50CDC37 | cell division cycle 37 homolog (S. cerevisiae) | IKK-beta interacts with Cdc37. This interaction was modeled on a demonstrated interaction between IKK-beta from an unspecified species and human Cdc37. | BIND | 11864612 |
| CFLAR | CASH | CASP8AP1 | CLARP | Casper | FLAME | FLAME-1 | FLAME1 | FLIP | I-FLICE | MRIT | c-FLIP | c-FLIPL | c-FLIPR | c-FLIPS | CASP8 and FADD-like apoptosis regulator | - | HPRD | 11002417 |
| CHUK | IKBKA | IKK-alpha | IKK1 | IKKA | NFKBIKA | TCF16 | conserved helix-loop-helix ubiquitous kinase | IKKbeta interacts with IKKalpha. | BIND | 15856023 |
| CHUK | IKBKA | IKK-alpha | IKK1 | IKKA | NFKBIKA | TCF16 | conserved helix-loop-helix ubiquitous kinase | IKK-alpha interacts with IKK-beta. | BIND | 9346485 |
| CHUK | IKBKA | IKK-alpha | IKK1 | IKKA | NFKBIKA | TCF16 | conserved helix-loop-helix ubiquitous kinase | - | HPRD,BioGRID | 9346241 |
| CTNNB1 | CTNNB | DKFZp686D02253 | FLJ25606 | FLJ37923 | catenin (cadherin-associated protein), beta 1, 88kDa | - | HPRD,BioGRID | 11527961 |
| EIF2AK2 | EIF2AK1 | MGC126524 | PKR | PRKR | eukaryotic translation initiation factor 2-alpha kinase 2 | - | HPRD,BioGRID | 10848580 |
| FANCA | FA | FA-H | FA1 | FAA | FACA | FAH | FANCH | MGC75158 | Fanconi anemia, complementation group A | - | HPRD,BioGRID | 12210728 |
| FOXO3 | AF6q21 | DKFZp781A0677 | FKHRL1 | FKHRL1P2 | FOXO2 | FOXO3A | MGC12739 | MGC31925 | forkhead box O3 | IKK-beta interacts with and phosphorylates FOXO3a. | BIND | 15084260 |
| HSP90AA1 | FLJ31884 | HSP86 | HSP89A | HSP90A | HSP90N | HSPC1 | HSPCA | HSPCAL1 | HSPCAL4 | HSPN | Hsp89 | Hsp90 | LAP2 | heat shock protein 90kDa alpha (cytosolic), class A member 1 | - | HPRD | 11864612 |
| HSPA8 | HSC54 | HSC70 | HSC71 | HSP71 | HSP73 | HSPA10 | LAP1 | MGC131511 | MGC29929 | NIP71 | heat shock 70kDa protein 8 | - | HPRD | 14743216 |
| IKBKAP | DKFZp781H1425 | DYS | ELP1 | FD | FLJ12497 | IKAP | IKI3 | TOT1 | inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase complex-associated protein | IKAP interacts with IKK-beta. | BIND | 9751059 |
| IKBKAP | DKFZp781H1425 | DYS | ELP1 | FD | FLJ12497 | IKAP | IKI3 | TOT1 | inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase complex-associated protein | - | HPRD,BioGRID | 9751059 |
| IKBKB | FLJ40509 | IKK-beta | IKK2 | IKKB | MGC131801 | NFKBIKB | inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase beta | IKK-beta interacts with another IKK-beta. | BIND | 9346485 |
| IKBKG | AMCBX1 | FIP-3 | FIP3 | Fip3p | IKK-gamma | IP | IP1 | IP2 | IPD2 | NEMO | inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma | - | HPRD,BioGRID | 10968790 |
| IKBKG | AMCBX1 | FIP-3 | FIP3 | Fip3p | IKK-gamma | IP | IP1 | IP2 | IPD2 | NEMO | inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma | NEMO interacts with IKK-beta. | BIND | 15802604 |
| IL8 | CXCL8 | GCP-1 | GCP1 | LECT | LUCT | LYNAP | MDNCF | MONAP | NAF | NAP-1 | NAP1 | interleukin 8 | The IL-8 promoter interacts with IKK-beta. | BIND | 15494311 |
| IRAK1 | IRAK | pelle | interleukin-1 receptor-associated kinase 1 | Affinity Capture-Western | BioGRID | 11096118 |
| IRS1 | HIRS-1 | insulin receptor substrate 1 | Affinity Capture-Western | BioGRID | 12351658 |
| MAP3K11 | MGC17114 | MLK-3 | MLK3 | PTK1 | SPRK | mitogen-activated protein kinase kinase kinase 11 | - | HPRD,BioGRID | 10713178 |
| MAP3K13 | LZK | MGC133196 | mitogen-activated protein kinase kinase kinase 13 | - | HPRD,BioGRID | 12492477 |
| MAP3K14 | FTDCR1B | HS | HSNIK | NIK | mitogen-activated protein kinase kinase kinase 14 | IKK-beta interacts with NIK. This interaction was modeled on a demonstrated interaction between human IKK-beta and NIK from an unspecified species. | BIND | 9346485 |
| MAP3K14 | FTDCR1B | HS | HSNIK | NIK | mitogen-activated protein kinase kinase kinase 14 | - | HPRD,BioGRID | 9346485 |
| NCOA3 | ACTR | AIB-1 | AIB1 | CAGH16 | CTG26 | KAT13B | MGC141848 | RAC3 | SRC3 | TNRC14 | TNRC16 | TRAM-1 | pCIP | nuclear receptor coactivator 3 | - | HPRD,BioGRID | 11971985 |
| NFKB1 | DKFZp686C01211 | EBP-1 | KBF1 | MGC54151 | NF-kappa-B | NFKB-p105 | NFKB-p50 | p105 | nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 | - | HPRD | 10469655|11158290 |
| NFKB1 | DKFZp686C01211 | EBP-1 | KBF1 | MGC54151 | NF-kappa-B | NFKB-p105 | NFKB-p50 | p105 | nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 | Affinity Capture-Western Biochemical Activity | BioGRID | 10469655 |11158290 |
| NFKBIA | IKBA | MAD-3 | NFKBI | nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha | IKK-beta interacts with and phosphorylates I-kappa-B-alpha. | BIND | 9346485 |
| NFKBIA | IKBA | MAD-3 | NFKBI | nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha | IKK-beta interacts with and phosphorylates I-kappa-B-alpha. This interaction was modelled on a demonstrated interaction between IKK-beta and I-kappa-B-alpha both from unspecified species. | BIND | 15808510 |
| NFKBIA | IKBA | MAD-3 | NFKBI | nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha | Reconstituted Complex | BioGRID | 9751059 |9891086 |
| NFKBIA | IKBA | MAD-3 | NFKBI | nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha | IKK-beta phosphorylates I-kappa-Ba. This interaction was modelled on a demonstrated interaction between IKK-beta from unknown species and I-kappa-Ba from unknown species. | BIND | 15084260 |
| NFKBIB | IKBB | TRIP9 | nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta | IKK-beta phosphorylates I-kappa-B-beta. | BIND | 9346485 |
| PPM1B | MGC21657 | PP2C-beta-X | PP2CB | PP2CBETA | PPC2BETAX | protein phosphatase 1B (formerly 2C), magnesium-dependent, beta isoform | Affinity Capture-Western Biochemical Activity | BioGRID | 14585847 |
| PRDX3 | AOP-1 | AOP1 | MER5 | MGC104387 | MGC24293 | PRO1748 | SP-22 | peroxiredoxin 3 | Affinity Capture-Western | BioGRID | 12492477 |
| PRKCQ | MGC126514 | MGC141919 | PRKCT | nPKC-theta | protein kinase C, theta | PKC-theta interacts with IKK-beta. | BIND | 15802604 |
| PRKCQ | MGC126514 | MGC141919 | PRKCT | nPKC-theta | protein kinase C, theta | - | HPRD,BioGRID | 11120819 |
| PRKDC | DNA-PKcs | DNAPK | DNPK1 | HYRC | HYRC1 | XRCC7 | p350 | protein kinase, DNA-activated, catalytic polypeptide | Biochemical Activity | BioGRID | 9632806 |
| TANK | I-TRAF | TRAF2 | TRAF family member-associated NFKB activator | - | HPRD | 12133833 |
| TNFAIP3 | A20 | MGC104522 | MGC138687 | MGC138688 | OTUD7C | TNFA1P2 | tumor necrosis factor, alpha-induced protein 3 | - | HPRD,BioGRID | 11594795 |
| TNFRSF1A | CD120a | FPF | MGC19588 | TBP1 | TNF-R | TNF-R-I | TNF-R55 | TNFAR | TNFR1 | TNFR55 | TNFR60 | p55 | p55-R | p60 | tumor necrosis factor receptor superfamily, member 1A | - | HPRD,BioGRID | 10755617 |
| TRAF2 | MGC:45012 | TRAP | TRAP3 | TNF receptor-associated factor 2 | - | HPRD,BioGRID | 11359906 |
| TRAF3IP2 | ACT1 | C6orf2 | C6orf4 | C6orf5 | C6orf6 | CIKS | DKFZp586G0522 | MGC3581 | TRAF3 interacting protein 2 | - | HPRD,BioGRID | 10962033 |
| TRPC4AP | C20orf188 | TRRP4AP | TRUSS | transient receptor potential cation channel, subfamily C, member 4 associated protein | Affinity Capture-Western | BioGRID | 14585990 |
| TUBA3C | TUBA2 | bA408E5.3 | tubulin, alpha 3c | - | HPRD | 14743216 |
| TUBB2B | DKFZp566F223 | FLJ98847 | MGC8685 | TUBB-PARALOG | bA506K6.1 | tubulin, beta 2B | - | HPRD | 14743216 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG MAPK SIGNALING PATHWAY | 267 | 205 | All SZGR 2.0 genes in this pathway |
| KEGG CHEMOKINE SIGNALING PATHWAY | 190 | 128 | All SZGR 2.0 genes in this pathway |
| KEGG APOPTOSIS | 88 | 62 | All SZGR 2.0 genes in this pathway |
| KEGG TOLL LIKE RECEPTOR SIGNALING PATHWAY | 102 | 88 | All SZGR 2.0 genes in this pathway |
| KEGG NOD LIKE RECEPTOR SIGNALING PATHWAY | 62 | 47 | All SZGR 2.0 genes in this pathway |
| KEGG RIG I LIKE RECEPTOR SIGNALING PATHWAY | 71 | 51 | All SZGR 2.0 genes in this pathway |
| KEGG CYTOSOLIC DNA SENSING PATHWAY | 56 | 44 | All SZGR 2.0 genes in this pathway |
| KEGG T CELL RECEPTOR SIGNALING PATHWAY | 108 | 89 | All SZGR 2.0 genes in this pathway |
| KEGG B CELL RECEPTOR SIGNALING PATHWAY | 75 | 56 | All SZGR 2.0 genes in this pathway |
| KEGG NEUROTROPHIN SIGNALING PATHWAY | 126 | 103 | All SZGR 2.0 genes in this pathway |
| KEGG INSULIN SIGNALING PATHWAY | 137 | 103 | All SZGR 2.0 genes in this pathway |
| KEGG ADIPOCYTOKINE SIGNALING PATHWAY | 67 | 57 | All SZGR 2.0 genes in this pathway |
| KEGG TYPE II DIABETES MELLITUS | 47 | 41 | All SZGR 2.0 genes in this pathway |
| KEGG EPITHELIAL CELL SIGNALING IN HELICOBACTER PYLORI INFECTION | 68 | 44 | All SZGR 2.0 genes in this pathway |
| KEGG PATHWAYS IN CANCER | 328 | 259 | All SZGR 2.0 genes in this pathway |
| KEGG PANCREATIC CANCER | 70 | 56 | All SZGR 2.0 genes in this pathway |
| KEGG PROSTATE CANCER | 89 | 75 | All SZGR 2.0 genes in this pathway |
| KEGG CHRONIC MYELOID LEUKEMIA | 73 | 59 | All SZGR 2.0 genes in this pathway |
| KEGG ACUTE MYELOID LEUKEMIA | 60 | 47 | All SZGR 2.0 genes in this pathway |
| KEGG SMALL CELL LUNG CANCER | 84 | 67 | All SZGR 2.0 genes in this pathway |
| BIOCARTA RELA PATHWAY | 16 | 12 | All SZGR 2.0 genes in this pathway |
| BIOCARTA AKT PATHWAY | 22 | 16 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CD40 PATHWAY | 15 | 13 | All SZGR 2.0 genes in this pathway |
| BIOCARTA TID PATHWAY | 19 | 15 | All SZGR 2.0 genes in this pathway |
| BIOCARTA RACCYCD PATHWAY | 26 | 23 | All SZGR 2.0 genes in this pathway |
| BIOCARTA KERATINOCYTE PATHWAY | 46 | 38 | All SZGR 2.0 genes in this pathway |
| BIOCARTA MAPK PATHWAY | 87 | 68 | All SZGR 2.0 genes in this pathway |
| BIOCARTA NTHI PATHWAY | 24 | 20 | All SZGR 2.0 genes in this pathway |
| BIOCARTA NFKB PATHWAY | 23 | 19 | All SZGR 2.0 genes in this pathway |
| BIOCARTA IL1R PATHWAY | 33 | 29 | All SZGR 2.0 genes in this pathway |
| BIOCARTA 41BB PATHWAY | 17 | 15 | All SZGR 2.0 genes in this pathway |
| BIOCARTA STRESS PATHWAY | 25 | 18 | All SZGR 2.0 genes in this pathway |
| BIOCARTA TNFR2 PATHWAY | 18 | 15 | All SZGR 2.0 genes in this pathway |
| BIOCARTA TOLL PATHWAY | 37 | 31 | All SZGR 2.0 genes in this pathway |
| PID FCER1 PATHWAY | 62 | 43 | All SZGR 2.0 genes in this pathway |
| PID BCR 5PATHWAY | 65 | 50 | All SZGR 2.0 genes in this pathway |
| PID GMCSF PATHWAY | 37 | 31 | All SZGR 2.0 genes in this pathway |
| PID NFKAPPAB ATYPICAL PATHWAY | 17 | 15 | All SZGR 2.0 genes in this pathway |
| PID TCR PATHWAY | 66 | 51 | All SZGR 2.0 genes in this pathway |
| PID NFKAPPAB CANONICAL PATHWAY | 23 | 20 | All SZGR 2.0 genes in this pathway |
| PID TRAIL PATHWAY | 28 | 20 | All SZGR 2.0 genes in this pathway |
| PID CD8 TCR PATHWAY | 53 | 42 | All SZGR 2.0 genes in this pathway |
| PID FAS PATHWAY | 38 | 29 | All SZGR 2.0 genes in this pathway |
| PID IL1 PATHWAY | 34 | 28 | All SZGR 2.0 genes in this pathway |
| PID MTOR 4PATHWAY | 69 | 55 | All SZGR 2.0 genes in this pathway |
| PID TNF PATHWAY | 46 | 33 | All SZGR 2.0 genes in this pathway |
| PID FOXO PATHWAY | 49 | 43 | All SZGR 2.0 genes in this pathway |
| PID P75 NTR PATHWAY | 69 | 51 | All SZGR 2.0 genes in this pathway |
| PID TAP63 PATHWAY | 54 | 40 | All SZGR 2.0 genes in this pathway |
| PID TOLL ENDOGENOUS PATHWAY | 25 | 19 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALLING BY NGF | 217 | 167 | All SZGR 2.0 genes in this pathway |
| REACTOME TRIF MEDIATED TLR3 SIGNALING | 74 | 54 | All SZGR 2.0 genes in this pathway |
| REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | 18 | 15 | All SZGR 2.0 genes in this pathway |
| REACTOME DOWNSTREAM SIGNALING EVENTS OF B CELL RECEPTOR BCR | 97 | 66 | All SZGR 2.0 genes in this pathway |
| REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | 64 | 43 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY THE B CELL RECEPTOR BCR | 126 | 90 | All SZGR 2.0 genes in this pathway |
| REACTOME TCR SIGNALING | 54 | 46 | All SZGR 2.0 genes in this pathway |
| REACTOME DOWNSTREAM TCR SIGNALING | 37 | 32 | All SZGR 2.0 genes in this pathway |
| REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | 12 | 8 | All SZGR 2.0 genes in this pathway |
| REACTOME P75NTR SIGNALS VIA NFKB | 14 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | 11 | 8 | All SZGR 2.0 genes in this pathway |
| REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | 81 | 61 | All SZGR 2.0 genes in this pathway |
| REACTOME TAK1 ACTIVATES NFKB BY PHOSPHORYLATION AND ACTIVATION OF IKKS COMPLEX | 23 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY ILS | 107 | 86 | All SZGR 2.0 genes in this pathway |
| REACTOME IL1 SIGNALING | 39 | 30 | All SZGR 2.0 genes in this pathway |
| REACTOME IRAK1 RECRUITS IKK COMPLEX | 10 | 8 | All SZGR 2.0 genes in this pathway |
| REACTOME TRAF6 MEDIATED NFKB ACTIVATION | 21 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME TRAF6 MEDIATED INDUCTION OF NFKB AND MAP KINASES UPON TLR7 8 OR 9 ACTIVATION | 77 | 57 | All SZGR 2.0 genes in this pathway |
| REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | 12 | 9 | All SZGR 2.0 genes in this pathway |
| REACTOME NFKB AND MAP KINASES ACTIVATION MEDIATED BY TLR4 SIGNALING REPERTOIRE | 72 | 53 | All SZGR 2.0 genes in this pathway |
| REACTOME RIG I MDA5 MEDIATED INDUCTION OF IFN ALPHA BETA PATHWAYS | 73 | 57 | All SZGR 2.0 genes in this pathway |
| REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | 10 | 7 | All SZGR 2.0 genes in this pathway |
| REACTOME MYD88 MAL CASCADE INITIATED ON PLASMA MEMBRANE | 83 | 63 | All SZGR 2.0 genes in this pathway |
| REACTOME INNATE IMMUNE SYSTEM | 279 | 178 | All SZGR 2.0 genes in this pathway |
| REACTOME ACTIVATED TLR4 SIGNALLING | 93 | 69 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME TOLL RECEPTOR CASCADES | 118 | 84 | All SZGR 2.0 genes in this pathway |
| REACTOME ADAPTIVE IMMUNE SYSTEM | 539 | 350 | All SZGR 2.0 genes in this pathway |
| REACTOME NOD1 2 SIGNALING PATHWAY | 30 | 22 | All SZGR 2.0 genes in this pathway |
| REACTOME CYTOKINE SIGNALING IN IMMUNE SYSTEM | 270 | 204 | All SZGR 2.0 genes in this pathway |
| REACTOME NUCLEOTIDE BINDING DOMAIN LEUCINE RICH REPEAT CONTAINING RECEPTOR NLR SIGNALING PATHWAYS | 46 | 33 | All SZGR 2.0 genes in this pathway |
| NAKAMURA TUMOR ZONE PERIPHERAL VS CENTRAL DN | 634 | 384 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 2 UP | 418 | 263 | All SZGR 2.0 genes in this pathway |
| ODONNELL TFRC TARGETS UP | 456 | 228 | All SZGR 2.0 genes in this pathway |
| ODONNELL TARGETS OF MYC AND TFRC UP | 83 | 50 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 3 DN | 229 | 142 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY SERUM DEPRIVATION UP | 552 | 347 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND SERUM DEPRIVATION UP | 211 | 136 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN DN | 770 | 415 | All SZGR 2.0 genes in this pathway |
| NUNODA RESPONSE TO DASATINIB IMATINIB DN | 13 | 10 | All SZGR 2.0 genes in this pathway |
| GILMORE CORE NFKB PATHWAY | 14 | 10 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER APOCRINE VS LUMINAL | 326 | 213 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| FERREIRA EWINGS SARCOMA UNSTABLE VS STABLE UP | 167 | 92 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 8P12 P11 AMPLICON | 57 | 32 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY MUTATED AND AMPLIFIED IN BREAST CANCER | 94 | 60 | All SZGR 2.0 genes in this pathway |
| BYSTROEM CORRELATED WITH IL5 UP | 51 | 30 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| RODWELL AGING KIDNEY UP | 487 | 303 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 5 | 482 | 296 | All SZGR 2.0 genes in this pathway |
| FINETTI BREAST CANCERS KINOME BLUE | 21 | 16 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER RELAPSE IN BRAIN DN | 85 | 58 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL DN | 701 | 446 | All SZGR 2.0 genes in this pathway |
| BONOME OVARIAN CANCER SURVIVAL SUBOPTIMAL DEBULKING | 510 | 309 | All SZGR 2.0 genes in this pathway |
| GRADE COLON CANCER UP | 871 | 505 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 UP | 430 | 288 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-199 | 267 | 274 | 1A,m8 | hsa-miR-199a | CCCAGUGUUCAGACUACCUGUUC |
| hsa-miR-199b | CCCAGUGUUUAGACUAUCUGUUC | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.