Gene Page: ITK
Summary ?
| GeneID | 3702 |
| Symbol | ITK |
| Synonyms | EMT|LPFS1|LYK|PSCTK2 |
| Description | IL2 inducible T-cell kinase |
| Reference | MIM:186973|HGNC:HGNC:6171|Ensembl:ENSG00000113263|HPRD:01746|Vega:OTTHUMG00000130245 |
| Gene type | protein-coding |
| Map location | 5q31-q32 |
| Pascal p-value | 0.055 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.0032 | |
| GSMA_IIE | Genome scan meta-analysis (European-ancestry samples) | Psr: 0.01718 | |
| GSMA_IIA | Genome scan meta-analysis (All samples) | Psr: 0.00459 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0385 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
General gene expression (GTEx)
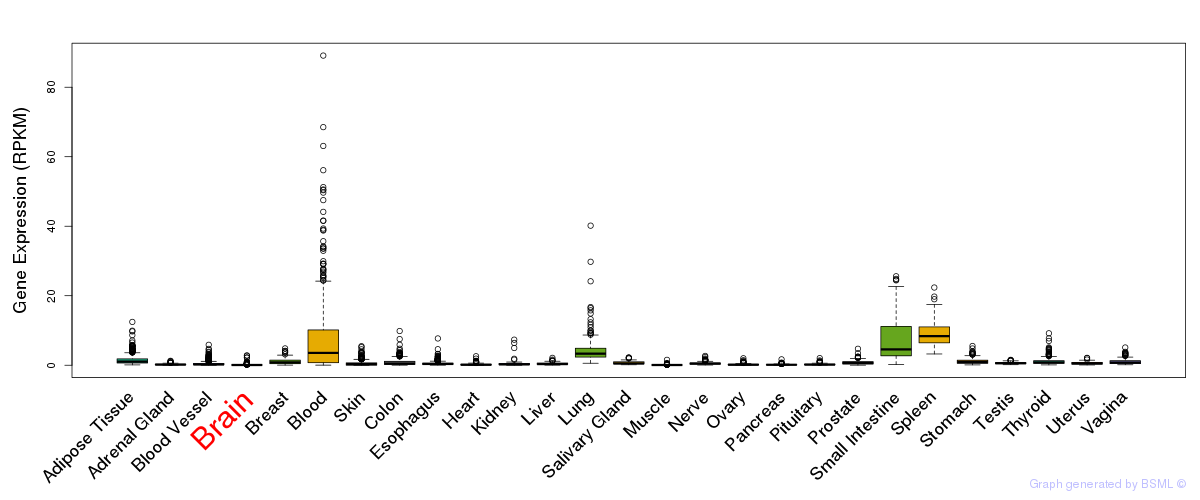
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| RAD23A | 0.76 | 0.79 |
| THOP1 | 0.74 | 0.77 |
| DVL1 | 0.74 | 0.75 |
| AKT1S1 | 0.74 | 0.74 |
| PIM3 | 0.74 | 0.79 |
| TFE3 | 0.73 | 0.73 |
| AL355312.3 | 0.73 | 0.78 |
| CXXC5 | 0.72 | 0.72 |
| MYPOP | 0.72 | 0.75 |
| AL355987.1 | 0.72 | 0.74 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.21 | -0.41 | -0.24 |
| FAM159B | -0.38 | -0.45 |
| AL050337.1 | -0.36 | -0.27 |
| AF347015.18 | -0.35 | -0.23 |
| C1orf54 | -0.35 | -0.22 |
| AC135724.1 | -0.34 | -0.30 |
| SYCP3 | -0.34 | -0.31 |
| CLEC2B | -0.34 | -0.19 |
| AF347015.8 | -0.33 | -0.22 |
| MT-ATP8 | -0.33 | -0.22 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0005515 | protein binding | IEA | - | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0004715 | non-membrane spanning protein tyrosine kinase activity | TAS | 8364206 | |
| GO:0016740 | transferase activity | IEA | - | |
| GO:0008270 | zinc ion binding | IEA | - | |
| GO:0016301 | kinase activity | IEA | - | |
| GO:0046872 | metal ion binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006468 | protein amino acid phosphorylation | IEA | - | |
| GO:0007242 | intracellular signaling cascade | IEA | - | |
| GO:0006968 | cellular defense response | TAS | 8364206 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005829 | cytosol | EXP | 11048639 |17652306 | |
| GO:0005886 | plasma membrane | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| CBL | C-CBL | CBL2 | RNF55 | Cas-Br-M (murine) ecotropic retroviral transforming sequence | - | HPRD,BioGRID | 8810341 |
| CD2 | SRBC | T11 | CD2 molecule | Affinity Capture-Western | BioGRID | 9677430 |
| CD28 | MGC138290 | Tp44 | CD28 molecule | - | HPRD | 10586033 |
| FYN | MGC45350 | SLK | SYN | FYN oncogene related to SRC, FGR, YES | - | HPRD,BioGRID | 8810341 |
| GRB2 | ASH | EGFRBP-GRB2 | Grb3-3 | MST084 | MSTP084 | growth factor receptor-bound protein 2 | - | HPRD,BioGRID | 10636929 |
| HNRNPK | CSBP | FLJ41122 | HNRPK | TUNP | heterogeneous nuclear ribonucleoprotein K | Itk interacts with hnRNPK. This interaction was modeled on a demonstrated interaction between mouse Itk and human hnRNPK. | BIND | 8810341 |
| HNRNPK | CSBP | FLJ41122 | HNRPK | TUNP | heterogeneous nuclear ribonucleoprotein K | - | HPRD,BioGRID | 8810341 |
| ITK | EMT | LYK | MGC126257 | MGC126258 | PSCTK2 | IL2-inducible T-cell kinase | Biochemical Activity | BioGRID | 12163161 |
| KHDRBS1 | FLJ34027 | Sam68 | p62 | KH domain containing, RNA binding, signal transduction associated 1 | - | HPRD,BioGRID | 8810341 |8985255 |
| KPNA1 | IPOA5 | NPI-1 | RCH2 | SRP1 | karyopherin alpha 1 (importin alpha 5) | - | HPRD | 11581171 |
| KPNA2 | IPOA1 | QIP2 | RCH1 | SRP1alpha | karyopherin alpha 2 (RAG cohort 1, importin alpha 1) | Affinity Capture-Western Reconstituted Complex | BioGRID | 11581171 |
| KPNA2 | IPOA1 | QIP2 | RCH1 | SRP1alpha | karyopherin alpha 2 (RAG cohort 1, importin alpha 1) | Itk interacts with and phosphorylates Rch1-alpha. | BIND | 11581171 |
| LAT | LAT1 | pp36 | linker for activation of T cells | - | HPRD | 10506192|12186560 |
| LAT | LAT1 | pp36 | linker for activation of T cells | Affinity Capture-Western Biochemical Activity | BioGRID | 10506192 |12186560 |
| LCP2 | SLP-76 | SLP76 | lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa) | - | HPRD,BioGRID | 10636929 |
| PLCG1 | PLC-II | PLC1 | PLC148 | PLCgamma1 | phospholipase C, gamma 1 | Affinity Capture-Western Biochemical Activity Reconstituted Complex | BioGRID | 10586033 |12163161 |
| PLCG1 | PLC-II | PLC1 | PLC148 | PLCgamma1 | phospholipase C, gamma 1 | - | HPRD | 10586033|12163161 |
| PPIA | CYPA | CYPH | MGC117158 | MGC12404 | MGC23397 | peptidylprolyl isomerase A (cyclophilin A) | - | HPRD,BioGRID | 11830645 |
| SOCS1 | CIS1 | CISH1 | JAB | SOCS-1 | SSI-1 | SSI1 | TIP3 | suppressor of cytokine signaling 1 | Socs1 interacts with Itk. This interaction was modeled on a demonstrated interaction between mouse Socs1 and Itk from an unspecified species. | BIND | 10022833 |
| SOCS1 | CIS1 | CISH1 | JAB | SOCS-1 | SSI-1 | SSI1 | TIP3 | suppressor of cytokine signaling 1 | - | HPRD,BioGRID | 10022833 |
| SRC | ASV | SRC1 | c-SRC | p60-Src | v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (avian) | Reconstituted Complex | BioGRID | 10636929 |
| WAS | IMD2 | THC | WASP | Wiskott-Aldrich syndrome (eczema-thrombocytopenia) | - | HPRD,BioGRID | 8810341 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CHEMOKINE SIGNALING PATHWAY | 190 | 128 | All SZGR 2.0 genes in this pathway |
| KEGG T CELL RECEPTOR SIGNALING PATHWAY | 108 | 89 | All SZGR 2.0 genes in this pathway |
| KEGG LEUKOCYTE TRANSENDOTHELIAL MIGRATION | 118 | 78 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CTLA4 PATHWAY | 21 | 18 | All SZGR 2.0 genes in this pathway |
| ST T CELL SIGNAL TRANSDUCTION | 45 | 33 | All SZGR 2.0 genes in this pathway |
| PID FCER1 PATHWAY | 62 | 43 | All SZGR 2.0 genes in this pathway |
| PID TCR PATHWAY | 66 | 51 | All SZGR 2.0 genes in this pathway |
| PID PI3KCI PATHWAY | 49 | 40 | All SZGR 2.0 genes in this pathway |
| REACTOME TCR SIGNALING | 54 | 46 | All SZGR 2.0 genes in this pathway |
| REACTOME GENERATION OF SECOND MESSENGER MOLECULES | 27 | 25 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME ADAPTIVE IMMUNE SYSTEM | 539 | 350 | All SZGR 2.0 genes in this pathway |
| JAATINEN HEMATOPOIETIC STEM CELL DN | 226 | 132 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 2B | 392 | 251 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER CLUSTER 1 | 44 | 23 | All SZGR 2.0 genes in this pathway |
| XU HGF TARGETS REPRESSED BY AKT1 DN | 95 | 58 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| KUMAR TARGETS OF MLL AF9 FUSION | 405 | 264 | All SZGR 2.0 genes in this pathway |
| SU THYMUS | 20 | 12 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 STIMULATED | 1022 | 619 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| MARSON FOXP3 TARGETS STIMULATED UP | 29 | 19 | All SZGR 2.0 genes in this pathway |
| MARSON FOXP3 CORE DIRECT TARGETS | 19 | 14 | All SZGR 2.0 genes in this pathway |
| KONDO PROSTATE CANCER WITH H3K27ME3 | 196 | 93 | All SZGR 2.0 genes in this pathway |
| FINETTI BREAST CANCER KINOME GREEN | 16 | 14 | All SZGR 2.0 genes in this pathway |
| WALLACE PROSTATE CANCER RACE UP | 299 | 167 | All SZGR 2.0 genes in this pathway |
| WORSCHECH TUMOR REJECTION UP | 56 | 32 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER LUMINAL B DN | 564 | 326 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER NORMAL LIKE UP | 476 | 285 | All SZGR 2.0 genes in this pathway |
| FIRESTEIN PROLIFERATION | 175 | 125 | All SZGR 2.0 genes in this pathway |
| GRESHOCK CANCER COPY NUMBER UP | 323 | 240 | All SZGR 2.0 genes in this pathway |
| BOYLAN MULTIPLE MYELOMA C D DN | 252 | 155 | All SZGR 2.0 genes in this pathway |
| LEE DIFFERENTIATING T LYMPHOCYTE | 200 | 115 | All SZGR 2.0 genes in this pathway |
| POOLA INVASIVE BREAST CANCER UP | 288 | 168 | All SZGR 2.0 genes in this pathway |
| MARSON FOXP3 TARGETS DN | 54 | 38 | All SZGR 2.0 genes in this pathway |
| BOYLAN MULTIPLE MYELOMA PCA1 UP | 101 | 66 | All SZGR 2.0 genes in this pathway |
| KASLER HDAC7 TARGETS 1 UP | 194 | 133 | All SZGR 2.0 genes in this pathway |
| BOSCO ALLERGEN INDUCED TH2 ASSOCIATED MODULE | 151 | 86 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-155 | 69 | 75 | m8 | hsa-miR-155 | UUAAUGCUAAUCGUGAUAGGGG |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.