Gene Page: KCNS3
Summary ?
| GeneID | 3790 |
| Symbol | KCNS3 |
| Synonyms | KV9.3 |
| Description | potassium voltage-gated channel modifier subfamily S member 3 |
| Reference | MIM:603888|HGNC:HGNC:6302|Ensembl:ENSG00000170745|HPRD:04865|Vega:OTTHUMG00000044150 |
| Gene type | protein-coding |
| Map location | 2p24 |
| Pascal p-value | 1.994E-5 |
| Fetal beta | -2.223 |
| DMG | 2 (# studies) |
| eGene | Caudate basal ganglia |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Nishioka_2013 | Genome-wide DNA methylation analysis | The authors investigated the methylation profiles of DNA in peripheral blood cells from 18 patients with first-episode schizophrenia (FESZ) and from 15 normal controls. | 2 |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 2 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Expression | Meta-analysis of gene expression | P value: 1.544 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg26119806 | 2 | 18061572 | KCNS3 | 3.38E-4 | -0.707 | 0.041 | DMG:Wockner_2014 |
| cg20940661 | 2 | 18060086 | KCNS3 | -0.022 | 0.46 | DMG:Nishioka_2013 |
Section II. Transcriptome annotation
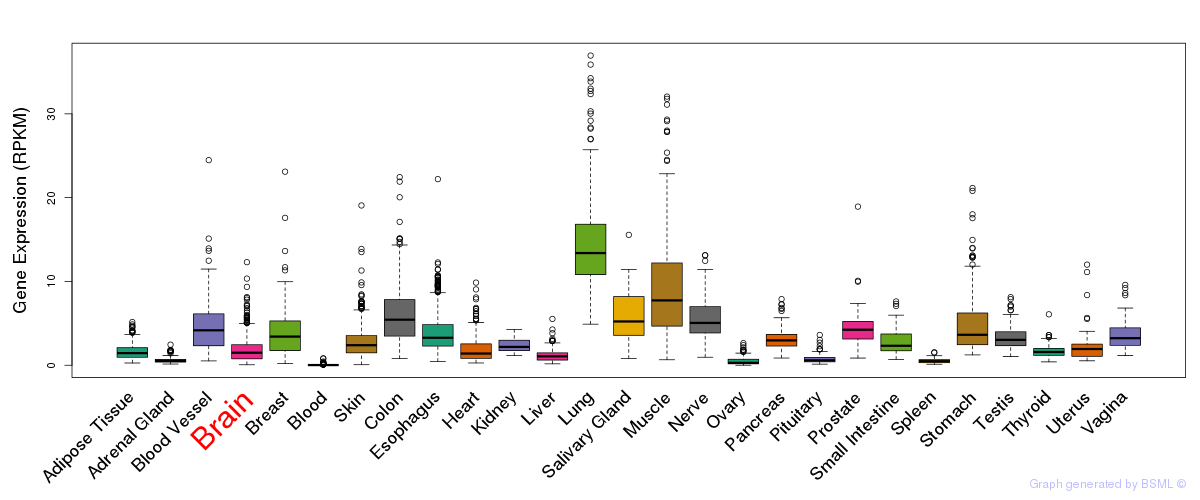
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| LRP12 | 0.82 | 0.90 |
| WDR47 | 0.82 | 0.90 |
| SMAP1 | 0.81 | 0.90 |
| PANK1 | 0.80 | 0.88 |
| PAPD5 | 0.80 | 0.90 |
| RALGPS2 | 0.80 | 0.88 |
| GFPT1 | 0.80 | 0.88 |
| TTC13 | 0.80 | 0.87 |
| VEZT | 0.80 | 0.89 |
| TRIM36 | 0.80 | 0.87 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MT-CO2 | -0.64 | -0.82 |
| AF347015.31 | -0.64 | -0.80 |
| AF347015.33 | -0.63 | -0.78 |
| FXYD1 | -0.62 | -0.79 |
| MT-CYB | -0.61 | -0.77 |
| HIGD1B | -0.61 | -0.79 |
| AF347015.27 | -0.60 | -0.76 |
| HLA-F | -0.60 | -0.69 |
| AF347015.8 | -0.60 | -0.78 |
| IFI27 | -0.60 | -0.76 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005515 | protein binding | IEA | - | |
| GO:0005244 | voltage-gated ion channel activity | IEA | - | |
| GO:0005251 | delayed rectifier potassium channel activity | TAS | 10484328 | |
| GO:0015459 | potassium channel regulator activity | TAS | 10484328 | |
| GO:0030955 | potassium ion binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006811 | ion transport | IEA | - | |
| GO:0006813 | potassium ion transport | TAS | 10484328 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0016021 | integral to membrane | IEA | - | |
| GO:0005886 | plasma membrane | IEA | - | |
| GO:0008076 | voltage-gated potassium channel complex | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| REACTOME NEURONAL SYSTEM | 279 | 221 | All SZGR 2.0 genes in this pathway |
| REACTOME INTEGRATION OF ENERGY METABOLISM | 120 | 84 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | 43 | 28 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF INSULIN SECRETION | 93 | 65 | All SZGR 2.0 genes in this pathway |
| REACTOME VOLTAGE GATED POTASSIUM CHANNELS | 43 | 32 | All SZGR 2.0 genes in this pathway |
| REACTOME POTASSIUM CHANNELS | 98 | 68 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA DN | 526 | 357 | All SZGR 2.0 genes in this pathway |
| DAVICIONI PAX FOXO1 SIGNATURE IN ARMS UP | 59 | 38 | All SZGR 2.0 genes in this pathway |
| IGARASHI ATF4 TARGETS DN | 90 | 65 | All SZGR 2.0 genes in this pathway |
| DAVICIONI TARGETS OF PAX FOXO1 FUSIONS UP | 255 | 177 | All SZGR 2.0 genes in this pathway |
| LIU CMYB TARGETS UP | 165 | 106 | All SZGR 2.0 genes in this pathway |
| LIU VMYB TARGETS UP | 127 | 78 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN NPM1 SIGNATURE 3 UP | 341 | 197 | All SZGR 2.0 genes in this pathway |
| OZEN MIR125B1 TARGETS | 25 | 15 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES 1 | 379 | 235 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| GEORGES TARGETS OF MIR192 AND MIR215 | 893 | 528 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 18HR UP | 178 | 111 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 3 | 720 | 440 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL CULTURED VS FRESH UP | 425 | 298 | All SZGR 2.0 genes in this pathway |
| SHEDDEN LUNG CANCER GOOD SURVIVAL A4 | 196 | 124 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3 | 1069 | 729 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3K4ME2 AND H3K27ME3 | 349 | 234 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN NPC HCP WITH H3K27ME3 | 341 | 243 | All SZGR 2.0 genes in this pathway |
| ZWANG EGF INTERVAL DN | 214 | 124 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-125/351 | 95 | 101 | m8 | hsa-miR-125bbrain | UCCCUGAGACCCUAACUUGUGA |
| hsa-miR-125abrain | UCCCUGAGACCCUUUAACCUGUG | ||||
| miR-135 | 112 | 118 | 1A | hsa-miR-135a | UAUGGCUUUUUAUUCCUAUGUGA |
| hsa-miR-135b | UAUGGCUUUUCAUUCCUAUGUG | ||||
| miR-142-5p | 56 | 62 | 1A | hsa-miR-142-5p | CAUAAAGUAGAAAGCACUAC |
| miR-320 | 53 | 60 | 1A,m8 | hsa-miR-320 | AAAAGCUGGGUUGAGAGGGCGAA |
| miR-33 | 400 | 406 | m8 | hsa-miR-33 | GUGCAUUGUAGUUGCAUUG |
| hsa-miR-33b | GUGCAUUGCUGUUGCAUUGCA | ||||
| miR-409-3p | 124 | 130 | 1A | hsa-miR-409-3p | CGAAUGUUGCUCGGUGAACCCCU |
| miR-496 | 377 | 383 | m8 | hsa-miR-496 | AUUACAUGGCCAAUCUC |
| hsa-miR-496 | AUUACAUGGCCAAUCUC | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.