Gene Page: NEDD4
Summary ?
| GeneID | 4734 |
| Symbol | NEDD4 |
| Synonyms | NEDD4-1|RPF1 |
| Description | neural precursor cell expressed, developmentally down-regulated 4, E3 ubiquitin protein ligase |
| Reference | MIM:602278|HGNC:HGNC:7727|Ensembl:ENSG00000069869|HPRD:03786|Vega:OTTHUMG00000132015 |
| Gene type | protein-coding |
| Map location | 15q |
| Pascal p-value | 0.031 |
| Fetal beta | 0.738 |
| Support | G2Cdb.humanPSD G2Cdb.humanPSP CompositeSet Darnell FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0132 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
General gene expression (GTEx)
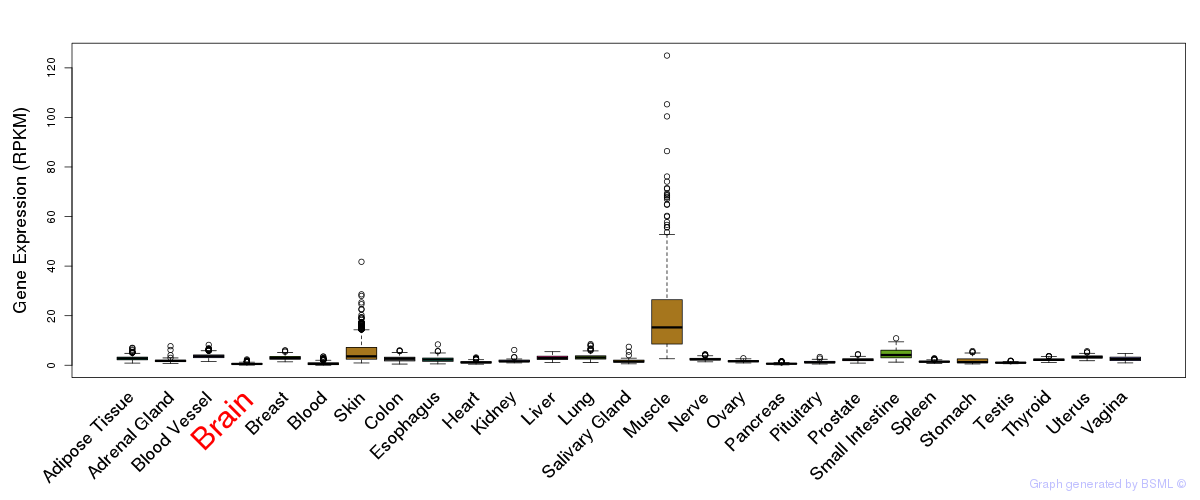
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| BOC | 0.73 | 0.53 |
| PDLIM1 | 0.67 | 0.40 |
| DCT | 0.67 | 0.00 |
| DPF3 | 0.66 | 0.38 |
| COL2A1 | 0.66 | 0.49 |
| HUNK | 0.65 | 0.78 |
| CDON | 0.64 | 0.75 |
| DUSP4 | 0.64 | 0.83 |
| TNFRSF19 | 0.64 | 0.67 |
| BHLHE22 | 0.63 | 0.85 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| C5orf53 | -0.41 | -0.60 |
| AIFM3 | -0.41 | -0.65 |
| PTH1R | -0.40 | -0.59 |
| HLA-F | -0.40 | -0.62 |
| AF347015.27 | -0.39 | -0.72 |
| MT-CO2 | -0.39 | -0.74 |
| AF347015.31 | -0.39 | -0.71 |
| AF347015.33 | -0.38 | -0.71 |
| IFI27 | -0.38 | -0.72 |
| LDHD | -0.38 | -0.51 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005515 | protein binding | IPI | 17719543 | |
| GO:0004842 | ubiquitin-protein ligase activity | IEA | - | |
| GO:0016874 | ligase activity | IEA | - | |
| GO:0016881 | acid-amino acid ligase activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006464 | protein modification process | IEA | - | |
| GO:0006511 | ubiquitin-dependent protein catabolic process | IEA | - | |
| GO:0044419 | interspecies interaction between organisms | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000151 | ubiquitin ligase complex | IEA | - | |
| GO:0005829 | cytosol | IEA | - | |
| GO:0005622 | intracellular | IEA | - | |
| GO:0005737 | cytoplasm | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ANXA13 | ANX13 | ISA | MGC150460 | annexin A13 | - | HPRD,BioGRID | 10871286 |
| CBLB | DKFZp686J10223 | DKFZp779A0729 | DKFZp779F1443 | FLJ36865 | FLJ41152 | Nbla00127 | RNF56 | Cas-Br-M (murine) ecotropic retroviral transforming sequence b | - | HPRD,BioGRID | 12907674 |
| CDC25C | CDC25 | cell division cycle 25 homolog C (S. pombe) | - | HPRD,BioGRID | 10037602 |
| DAZAP2 | KIAA0058 | MGC14319 | MGC766 | PRTB | DAZ associated protein 2 | - | HPRD | 11342538 |
| EPS15 | AF-1P | AF1P | MLLT5 | epidermal growth factor receptor pathway substrate 15 | - | HPRD,BioGRID | 11919637 |
| GRB10 | GRB-IR | Grb-10 | IRBP | KIAA0207 | MEG1 | RSS | growth factor receptor-bound protein 10 | - | HPRD | 12697834 |
| GRB10 | GRB-IR | Grb-10 | IRBP | KIAA0207 | MEG1 | RSS | growth factor receptor-bound protein 10 | - | HPRD | 10446181 |12697834|12697834 |
| HGS | HRS | ZFYVE8 | hepatocyte growth factor-regulated tyrosine kinase substrate | - | HPRD,BioGRID | 12230472 |
| IGF1R | CD221 | IGFIR | JTK13 | MGC142170 | MGC142172 | MGC18216 | insulin-like growth factor 1 receptor | - | HPRD,BioGRID | 12697834 |
| LITAF | FLJ38636 | MGC116698 | MGC116700 | MGC116701 | MGC125274 | MGC125275 | MGC125276 | PIG7 | SIMPLE | TP53I7 | lipopolysaccharide-induced TNF factor | - | HPRD | 11042109 |
| N4BP2 | B3BP | FLJ10680 | KIAA1413 | NEDD4 binding protein 2 | - | HPRD | 11717310 |
| N4BP3 | KIAA0341 | Nedd4 binding protein 3 | - | HPRD,BioGRID | 11717310 |
| NDFIP1 | MGC10924 | N4WBP5 | Nedd4 family interacting protein 1 | - | HPRD | 11042109|11748237 |
| NDFIP2 | FLJ25842 | KIAA1165 | N4wbp5a | Nedd4 family interacting protein 2 | - | HPRD | 12050153 |12796489 |15252135|12796489 |
| NDFIP2 | FLJ25842 | KIAA1165 | N4wbp5a | Nedd4 family interacting protein 2 | - | HPRD,BioGRID | 12050153 |12796489 |15252135 |
| NFE2 | NF-E2 | p45 | nuclear factor (erythroid-derived 2), 45kDa | p45 interacts with NEDD4. This interaction was modeled on a demonstrated interaction between human p45 and mouse NEDD4. | BIND | 9305852 |
| PMEPA1 | STAG1 | TMEPAI | prostate transmembrane protein, androgen induced 1 | - | HPRD,BioGRID | 11042109 |12907594 |
| PRRG2 | PRGP2 | proline rich Gla (G-carboxyglutamic acid) 2 | - | HPRD | 11042109 |
| SCNN1A | ENaCa | ENaCalpha | FLJ21883 | SCNEA | SCNN1 | sodium channel, nonvoltage-gated 1 alpha | - | HPRD,BioGRID | 10642508 |
| SCNN1A | ENaCa | ENaCalpha | FLJ21883 | SCNEA | SCNN1 | sodium channel, nonvoltage-gated 1 alpha | - | HPRD | 10212229 |10642508 |11359767|10642508 |
| SCNN1B | ENaCb | ENaCbeta | SCNEB | sodium channel, nonvoltage-gated 1, beta | - | HPRD | 10212229 |10642508 |11359767|10642508 |
| SCNN1B | ENaCb | ENaCbeta | SCNEB | sodium channel, nonvoltage-gated 1, beta | - | HPRD,BioGRID | 10642508 |
| SCNN1G | ENaCg | ENaCgamma | PHA1 | SCNEG | sodium channel, nonvoltage-gated 1, gamma | - | HPRD,BioGRID | 10642508 |
| SCNN1G | ENaCg | ENaCgamma | PHA1 | SCNEG | sodium channel, nonvoltage-gated 1, gamma | - | HPRD | 10212229 |10642508 |11359767|10642508 |
| SGK1 | SGK | serum/glucocorticoid regulated kinase 1 | - | HPRD,BioGRID | 11696533 |
| SH3KBP1 | CIN85 | GIG10 | MIG18 | SH3-domain kinase binding protein 1 | - | HPRD,BioGRID | 12218189 |
| TP73 | P73 | tumor protein p73 | p73-alpha interacts with an unspecified isoform of Nedd4. | BIND | 15678106 |
| UBE2D2 | E2(17)KB2 | PUBC1 | UBC4 | UBC4/5 | UBCH5B | ubiquitin-conjugating enzyme E2D 2 (UBC4/5 homolog, yeast) | - | HPRD,BioGRID | 9182527 |
| UBE2D3 | E2(17)KB3 | MGC43926 | MGC5416 | UBC4/5 | UBCH5C | ubiquitin-conjugating enzyme E2D 3 (UBC4/5 homolog, yeast) | - | HPRD,BioGRID | 9990509 |
| UBE2E1 | UBCH6 | ubiquitin-conjugating enzyme E2E 1 (UBC4/5 homolog, yeast) | - | HPRD,BioGRID | 9990509 |
| UBE2L3 | E2-F1 | L-UBC | UBCH7 | UbcM4 | ubiquitin-conjugating enzyme E2L 3 | - | HPRD,BioGRID | 9990509 |
| WBP1 | MGC15305 | WBP-1 | WW domain binding protein 1 | - | HPRD | 11042109 |
| WBP2 | MGC18269 | WBP-2 | WW domain binding protein 2 | - | HPRD | 11042109 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG UBIQUITIN MEDIATED PROTEOLYSIS | 138 | 98 | All SZGR 2.0 genes in this pathway |
| KEGG ENDOCYTOSIS | 183 | 132 | All SZGR 2.0 genes in this pathway |
| PID ERBB4 PATHWAY | 38 | 32 | All SZGR 2.0 genes in this pathway |
| PID PS1 PATHWAY | 46 | 39 | All SZGR 2.0 genes in this pathway |
| PID VEGFR1 2 PATHWAY | 69 | 57 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY ERBB4 | 90 | 67 | All SZGR 2.0 genes in this pathway |
| REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | 66 | 45 | All SZGR 2.0 genes in this pathway |
| REACTOME INTERFERON SIGNALING | 159 | 116 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME ADAPTIVE IMMUNE SYSTEM | 539 | 350 | All SZGR 2.0 genes in this pathway |
| REACTOME CYTOKINE SIGNALING IN IMMUNE SYSTEM | 270 | 204 | All SZGR 2.0 genes in this pathway |
| REACTOME CLASS I MHC MEDIATED ANTIGEN PROCESSING PRESENTATION | 251 | 156 | All SZGR 2.0 genes in this pathway |
| REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | 212 | 129 | All SZGR 2.0 genes in this pathway |
| HUTTMANN B CLL POOR SURVIVAL UP | 276 | 187 | All SZGR 2.0 genes in this pathway |
| DEURIG T CELL PROLYMPHOCYTIC LEUKEMIA UP | 368 | 234 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 2 DN | 281 | 186 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION UP | 1278 | 748 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED UP | 633 | 376 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| RASHI RESPONSE TO IONIZING RADIATION 5 | 147 | 89 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER APOCRINE VS BASAL | 330 | 217 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 COMMON DN | 483 | 336 | All SZGR 2.0 genes in this pathway |
| GROSS HYPOXIA VIA ELK3 UP | 209 | 139 | All SZGR 2.0 genes in this pathway |
| WEI MYCN TARGETS WITH E BOX | 795 | 478 | All SZGR 2.0 genes in this pathway |
| RICKMAN TUMOR DIFFERENTIATED WELL VS MODERATELY DN | 110 | 64 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 6HR DN | 514 | 330 | All SZGR 2.0 genes in this pathway |
| PARK HSC VS MULTIPOTENT PROGENITORS UP | 19 | 14 | All SZGR 2.0 genes in this pathway |
| SASAKI ADULT T CELL LEUKEMIA | 176 | 122 | All SZGR 2.0 genes in this pathway |
| HESS TARGETS OF HOXA9 AND MEIS1 UP | 65 | 44 | All SZGR 2.0 genes in this pathway |
| VERHAAK AML WITH NPM1 MUTATED DN | 246 | 180 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR UP | 953 | 554 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR UP | 783 | 442 | All SZGR 2.0 genes in this pathway |
| GAVIN FOXP3 TARGETS CLUSTER P6 | 91 | 44 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS UP | 673 | 430 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS UP | 602 | 364 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS UP | 601 | 369 | All SZGR 2.0 genes in this pathway |
| TOYOTA TARGETS OF MIR34B AND MIR34C | 463 | 262 | All SZGR 2.0 genes in this pathway |
| VANASSE BCL2 TARGETS DN | 74 | 50 | All SZGR 2.0 genes in this pathway |
| HOFFMANN LARGE TO SMALL PRE BII LYMPHOCYTE UP | 163 | 102 | All SZGR 2.0 genes in this pathway |
| HOFFMANN SMALL PRE BII TO IMMATURE B LYMPHOCYTE UP | 70 | 49 | All SZGR 2.0 genes in this pathway |
| HOFFMANN IMMATURE TO MATURE B LYMPHOCYTE DN | 50 | 36 | All SZGR 2.0 genes in this pathway |
| HAN SATB1 TARGETS DN | 442 | 275 | All SZGR 2.0 genes in this pathway |
| BOYAULT LIVER CANCER SUBCLASS G56 UP | 12 | 9 | All SZGR 2.0 genes in this pathway |
| KYNG WERNER SYNDROM AND NORMAL AGING DN | 225 | 124 | All SZGR 2.0 genes in this pathway |
| ZHAN MULTIPLE MYELOMA CD2 DN | 46 | 29 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE LATE | 1137 | 655 | All SZGR 2.0 genes in this pathway |
| BILANGES SERUM SENSITIVE VIA TSC2 | 39 | 25 | All SZGR 2.0 genes in this pathway |
| KOINUMA TARGETS OF SMAD2 OR SMAD3 | 824 | 528 | All SZGR 2.0 genes in this pathway |
| IKEDA MIR30 TARGETS UP | 116 | 87 | All SZGR 2.0 genes in this pathway |
| YANG BCL3 TARGETS UP | 364 | 236 | All SZGR 2.0 genes in this pathway |
| KRIEG HYPOXIA NOT VIA KDM3A | 770 | 480 | All SZGR 2.0 genes in this pathway |
| FORTSCHEGGER PHF8 TARGETS DN | 784 | 464 | All SZGR 2.0 genes in this pathway |
| RAO BOUND BY SALL4 ISOFORM A | 182 | 108 | All SZGR 2.0 genes in this pathway |
| ZWANG CLASS 1 TRANSIENTLY INDUCED BY EGF | 516 | 308 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-128 | 381 | 387 | 1A | hsa-miR-128a | UCACAGUGAACCGGUCUCUUUU |
| hsa-miR-128b | UCACAGUGAACCGGUCUCUUUC | ||||
| miR-144 | 1112 | 1118 | m8 | hsa-miR-144 | UACAGUAUAGAUGAUGUACUAG |
| miR-27 | 381 | 388 | 1A,m8 | hsa-miR-27abrain | UUCACAGUGGCUAAGUUCCGC |
| hsa-miR-27bbrain | UUCACAGUGGCUAAGUUCUGC | ||||
| miR-30-5p | 942 | 949 | 1A,m8 | hsa-miR-30a-5p | UGUAAACAUCCUCGACUGGAAG |
| hsa-miR-30cbrain | UGUAAACAUCCUACACUCUCAGC | ||||
| hsa-miR-30dSZ | UGUAAACAUCCCCGACUGGAAG | ||||
| hsa-miR-30bSZ | UGUAAACAUCCUACACUCAGCU | ||||
| hsa-miR-30e-5p | UGUAAACAUCCUUGACUGGA | ||||
| hsa-miR-30a-5p | UGUAAACAUCCUCGACUGGAAG | ||||
| hsa-miR-30cbrain | UGUAAACAUCCUACACUCUCAGC | ||||
| hsa-miR-30dSZ | UGUAAACAUCCCCGACUGGAAG | ||||
| hsa-miR-30bSZ | UGUAAACAUCCUACACUCAGCU | ||||
| hsa-miR-30e-5p | UGUAAACAUCCUUGACUGGA | ||||
| miR-376 | 1122 | 1128 | 1A | hsa-miR-376a | AUCAUAGAGGAAAAUCCACGU |
| hsa-miR-376b | AUCAUAGAGGAAAAUCCAUGUU | ||||
| miR-9 | 1048 | 1055 | 1A,m8 | hsa-miR-9SZ | UCUUUGGUUAUCUAGCUGUAUGA |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.