Gene Page: NEUROD1
Summary ?
| GeneID | 4760 |
| Symbol | NEUROD1 |
| Synonyms | BETA2|BHF-1|MODY6|NEUROD|bHLHa3 |
| Description | neuronal differentiation 1 |
| Reference | MIM:601724|HGNC:HGNC:7762|Ensembl:ENSG00000162992|HPRD:03428|Vega:OTTHUMG00000132583 |
| Gene type | protein-coding |
| Map location | 2q32 |
| Pascal p-value | 0.323 |
| Fetal beta | 2.508 |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 2 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0024 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs16829545 | chr2 | 151977407 | NEUROD1 | 4760 | 1.658E-14 | trans | ||
| rs7584986 | chr2 | 184111432 | NEUROD1 | 4760 | 6.239E-6 | trans | ||
| rs7609691 | chr3 | 143427927 | NEUROD1 | 4760 | 0.2 | trans | ||
| rs2183142 | chr4 | 159232695 | NEUROD1 | 4760 | 0.01 | trans | ||
| rs10491487 | chr5 | 80323367 | NEUROD1 | 4760 | 0.02 | trans | ||
| rs6996695 | chr8 | 77540580 | NEUROD1 | 4760 | 0.06 | trans | ||
| rs11139116 | chr9 | 83896951 | NEUROD1 | 4760 | 0 | trans | ||
| rs12098525 | chr10 | 87501764 | NEUROD1 | 4760 | 0.12 | trans | ||
| rs16955618 | chr15 | 29937543 | NEUROD1 | 4760 | 1.335E-12 | trans | ||
| rs16945437 | chr17 | 11797650 | NEUROD1 | 4760 | 0.07 | trans |
Section II. Transcriptome annotation
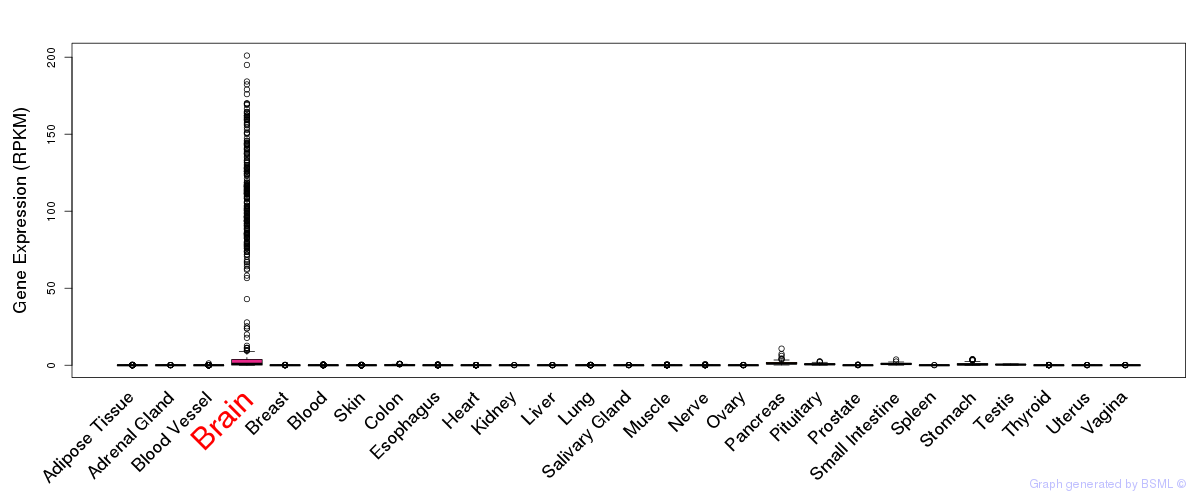
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| TRIP6 | 0.79 | 0.78 |
| SYDE1 | 0.76 | 0.75 |
| TRIP10 | 0.76 | 0.76 |
| INPPL1 | 0.76 | 0.76 |
| FKBP10 | 0.75 | 0.75 |
| MOV10 | 0.75 | 0.77 |
| LAMB2 | 0.74 | 0.76 |
| CTDSP1 | 0.74 | 0.76 |
| SIX5 | 0.73 | 0.75 |
| TEAD3 | 0.72 | 0.71 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| RALYL | -0.55 | -0.59 |
| PPP3CB | -0.53 | -0.56 |
| HMGCLL1 | -0.51 | -0.57 |
| PRKACB | -0.50 | -0.55 |
| FBXW7 | -0.50 | -0.54 |
| RGS7 | -0.50 | -0.49 |
| RAP1GDS1 | -0.49 | -0.53 |
| C20orf7 | -0.49 | -0.58 |
| ELOVL4 | -0.49 | -0.53 |
| C1orf52 | -0.49 | -0.51 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003700 | transcription factor activity | IDA | 10545951 | |
| GO:0003702 | RNA polymerase II transcription factor activity | IDA | 10545951 | |
| GO:0008134 | transcription factor binding | IPI | 10545951 | |
| GO:0016563 | transcription activator activity | IEA | - | |
| GO:0046982 | protein heterodimerization activity | IPI | 10545951 | |
| GO:0043565 | sequence-specific DNA binding | IDA | 10545951 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0022008 | neurogenesis | TAS | neurogenesis (GO term level: 6) | 8786144 |
| GO:0030902 | hindbrain development | IEA | Brain (GO term level: 8) | - |
| GO:0006355 | regulation of transcription, DNA-dependent | IEA | - | |
| GO:0007263 | nitric oxide mediated signal transduction | IDA | 17941991 | |
| GO:0009952 | anterior/posterior pattern formation | IEA | - | |
| GO:0009749 | response to glucose stimulus | IMP | 10545951 | |
| GO:0007275 | multicellular organismal development | IEA | - | |
| GO:0043066 | negative regulation of apoptosis | ISS | 9308961 | |
| GO:0031018 | endocrine pancreas development | ISS | 9308961 | |
| GO:0042593 | glucose homeostasis | ISS | 9308961 | |
| GO:0043010 | camera-type eye development | IEA | - | |
| GO:0045165 | cell fate commitment | IEA | - | |
| GO:0045944 | positive regulation of transcription from RNA polymerase II promoter | IEA | - | |
| GO:0048562 | embryonic organ morphogenesis | ISS | 9308961 | |
| GO:0050796 | regulation of insulin secretion | IC | 10545951 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005737 | cytoplasm | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| CALM1 | CALML2 | CAMI | DD132 | PHKD | calmodulin 1 (phosphorylase kinase, delta) | - | HPRD | 10757985 |
| CALM2 | CAMII | PHKD | PHKD2 | calmodulin 2 (phosphorylase kinase, delta) | - | HPRD | 10757985 |
| CALM3 | PHKD | PHKD3 | calmodulin 3 (phosphorylase kinase, delta) | - | HPRD | 10757985 |
| CCND1 | BCL1 | D11S287E | PRAD1 | U21B31 | cyclin D1 | Affinity Capture-Western Phenotypic Suppression | BioGRID | 11788592 |
| EP300 | KAT3B | p300 | E1A binding protein p300 | - | HPRD,BioGRID | 10545951 |
| HAP1 | HAP2 | HIP5 | HLP | hHLP1 | huntingtin-associated protein 1 | - | HPRD,BioGRID | 12881483 |
| HTT | HD | IT15 | huntingtin | Affinity Capture-Western | BioGRID | 12881483 |
| MAFA | RIPE3b1 | hMafA | v-maf musculoaponeurotic fibrosarcoma oncogene homolog A (avian) | - | HPRD,BioGRID | 15665000 |
| MAP3K10 | MLK2 | MST | mitogen-activated protein kinase kinase kinase 10 | Affinity Capture-Western Biochemical Activity Two-hybrid | BioGRID | 12881483 |
| PDX1 | GSF | IDX-1 | IPF1 | IUF1 | MODY4 | PDX-1 | STF-1 | pancreatic and duodenal homeobox 1 | - | HPRD,BioGRID | 10636926 |
| PITX1 | BFT | POTX | PTX1 | paired-like homeodomain 1 | - | HPRD | 9343431 |
| RREB1 | FINB | HNT | LZ321 | RREB-1 | Zep-1 | ras responsive element binding protein 1 | - | HPRD | 12482979 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG MATURITY ONSET DIABETES OF THE YOUNG | 25 | 19 | All SZGR 2.0 genes in this pathway |
| REACTOME DEVELOPMENTAL BIOLOGY | 396 | 292 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF BETA CELL DEVELOPMENT | 30 | 23 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | 20 | 15 | All SZGR 2.0 genes in this pathway |
| WILCOX RESPONSE TO PROGESTERONE UP | 152 | 90 | All SZGR 2.0 genes in this pathway |
| SABATES COLORECTAL ADENOMA DN | 291 | 176 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS C UP | 170 | 114 | All SZGR 2.0 genes in this pathway |
| RIZ ERYTHROID DIFFERENTIATION 6HR | 40 | 23 | All SZGR 2.0 genes in this pathway |
| HERNANDEZ ABERRANT MITOSIS BY DOCETACEL 4NM UP | 23 | 15 | All SZGR 2.0 genes in this pathway |
| MARTORIATI MDM4 TARGETS NEUROEPITHELIUM DN | 164 | 111 | All SZGR 2.0 genes in this pathway |
| BENPORATH SUZ12 TARGETS | 1038 | 678 | All SZGR 2.0 genes in this pathway |
| BENPORATH EED TARGETS | 1062 | 725 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| BENPORATH PRC2 TARGETS | 652 | 441 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 14HR DN | 298 | 200 | All SZGR 2.0 genes in this pathway |
| ZHOU PANCREATIC ENDOCRINE PROGENITOR | 14 | 11 | All SZGR 2.0 genes in this pathway |
| ZHOU PANCREATIC BETA CELL | 11 | 9 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3 | 1069 | 729 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MCV6 HCP WITH H3K27ME3 | 435 | 318 | All SZGR 2.0 genes in this pathway |
| SETLUR PROSTATE CANCER TMPRSS2 ERG FUSION DN | 20 | 12 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MEF HCP WITH H3K27ME3 | 590 | 403 | All SZGR 2.0 genes in this pathway |
| CARD MIR302A TARGETS | 77 | 62 | All SZGR 2.0 genes in this pathway |
| VANDESLUIS COMMD1 TARGETS GROUP 4 DN | 16 | 11 | All SZGR 2.0 genes in this pathway |
| MADAN DPPA4 TARGETS | 46 | 26 | All SZGR 2.0 genes in this pathway |
| ROLEF GLIS3 TARGETS | 37 | 29 | All SZGR 2.0 genes in this pathway |
| RAO BOUND BY SALL4 ISOFORM B | 517 | 302 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-101 | 1229 | 1236 | 1A,m8 | hsa-miR-101 | UACAGUACUGUGAUAACUGAAG |
| miR-103/107 | 90 | 96 | 1A | hsa-miR-103brain | AGCAGCAUUGUACAGGGCUAUGA |
| hsa-miR-107brain | AGCAGCAUUGUACAGGGCUAUCA | ||||
| miR-124.1 | 613 | 619 | m8 | hsa-miR-124a | UUAAGGCACGCGGUGAAUGCCA |
| miR-124/506 | 613 | 619 | 1A | hsa-miR-506 | UAAGGCACCCUUCUGAGUAGA |
| hsa-miR-124brain | UAAGGCACGCGGUGAAUGCC | ||||
| miR-128 | 36 | 42 | m8 | hsa-miR-128a | UCACAGUGAACCGGUCUCUUUU |
| hsa-miR-128b | UCACAGUGAACCGGUCUCUUUC | ||||
| miR-130/301 | 898 | 904 | m8 | hsa-miR-130abrain | CAGUGCAAUGUUAAAAGGGCAU |
| hsa-miR-301 | CAGUGCAAUAGUAUUGUCAAAGC | ||||
| hsa-miR-130bbrain | CAGUGCAAUGAUGAAAGGGCAU | ||||
| hsa-miR-454-3p | UAGUGCAAUAUUGCUUAUAGGGUUU | ||||
| miR-137 | 318 | 325 | 1A,m8 | hsa-miR-137 | UAUUGCUUAAGAAUACGCGUAG |
| miR-138 | 394 | 400 | 1A | hsa-miR-138brain | AGCUGGUGUUGUGAAUC |
| hsa-miR-138brain | AGCUGGUGUUGUGAAUC | ||||
| miR-144 | 1230 | 1236 | 1A | hsa-miR-144 | UACAGUAUAGAUGAUGUACUAG |
| miR-145 | 1215 | 1221 | 1A | hsa-miR-145 | GUCCAGUUUUCCCAGGAAUCCCUU |
| miR-153 | 334 | 341 | 1A,m8 | hsa-miR-153 | UUGCAUAGUCACAAAAGUGA |
| miR-17-5p/20/93.mr/106/519.d | 701 | 707 | 1A | hsa-miR-17-5p | CAAAGUGCUUACAGUGCAGGUAGU |
| hsa-miR-20abrain | UAAAGUGCUUAUAGUGCAGGUAG | ||||
| hsa-miR-106a | AAAAGUGCUUACAGUGCAGGUAGC | ||||
| hsa-miR-106bSZ | UAAAGUGCUGACAGUGCAGAU | ||||
| hsa-miR-20bSZ | CAAAGUGCUCAUAGUGCAGGUAG | ||||
| hsa-miR-519d | CAAAGUGCCUCCCUUUAGAGUGU | ||||
| miR-18 | 183 | 189 | 1A | hsa-miR-18a | UAAGGUGCAUCUAGUGCAGAUA |
| hsa-miR-18b | UAAGGUGCAUCUAGUGCAGUUA | ||||
| miR-19 | 897 | 903 | m8 | hsa-miR-19a | UGUGCAAAUCUAUGCAAAACUGA |
| hsa-miR-19b | UGUGCAAAUCCAUGCAAAACUGA | ||||
| hsa-miR-19a | UGUGCAAAUCUAUGCAAAACUGA | ||||
| hsa-miR-19b | UGUGCAAAUCCAUGCAAAACUGA | ||||
| hsa-miR-19a | UGUGCAAAUCUAUGCAAAACUGA | ||||
| hsa-miR-19b | UGUGCAAAUCCAUGCAAAACUGA | ||||
| miR-190 | 523 | 530 | 1A,m8 | hsa-miR-190 | UGAUAUGUUUGAUAUAUUAGGU |
| miR-217 | 1171 | 1177 | m8 | hsa-miR-217 | UACUGCAUCAGGAACUGAUUGGAU |
| miR-24 | 1046 | 1052 | m8 | hsa-miR-24SZ | UGGCUCAGUUCAGCAGGAACAG |
| miR-25/32/92/363/367 | 1298 | 1304 | 1A | hsa-miR-25brain | CAUUGCACUUGUCUCGGUCUGA |
| hsa-miR-32 | UAUUGCACAUUACUAAGUUGC | ||||
| hsa-miR-92 | UAUUGCACUUGUCCCGGCCUG | ||||
| hsa-miR-367 | AAUUGCACUUUAGCAAUGGUGA | ||||
| hsa-miR-92bSZ | UAUUGCACUCGUCCCGGCCUC | ||||
| miR-299-5p | 577 | 583 | 1A | hsa-miR-299-5p | UGGUUUACCGUCCCACAUACAU |
| miR-30-5p | 59 | 66 | 1A,m8 | hsa-miR-30a-5p | UGUAAACAUCCUCGACUGGAAG |
| hsa-miR-30cbrain | UGUAAACAUCCUACACUCUCAGC | ||||
| hsa-miR-30dSZ | UGUAAACAUCCCCGACUGGAAG | ||||
| hsa-miR-30bSZ | UGUAAACAUCCUACACUCAGCU | ||||
| hsa-miR-30e-5p | UGUAAACAUCCUUGACUGGA | ||||
| miR-328 | 358 | 364 | 1A | hsa-miR-328brain | CUGGCCCUCUCUGCCCUUCCGU |
| miR-331 | 162 | 168 | 1A | hsa-miR-331brain | GCCCCUGGGCCUAUCCUAGAA |
| miR-34b | 145 | 151 | m8 | hsa-miR-34b | UAGGCAGUGUCAUUAGCUGAUUG |
| miR-363 | 99 | 105 | 1A | hsa-miR-363 | AUUGCACGGUAUCCAUCUGUAA |
| miR-380-5p | 391 | 397 | 1A | hsa-miR-380-5p | UGGUUGACCAUAGAACAUGCGC |
| hsa-miR-563 | AGGUUGACAUACGUUUCCC | ||||
| miR-383 | 295 | 301 | m8 | hsa-miR-383brain | AGAUCAGAAGGUGAUUGUGGCU |
| miR-448 | 335 | 341 | 1A | hsa-miR-448 | UUGCAUAUGUAGGAUGUCCCAU |
| miR-494 | 967 | 973 | m8 | hsa-miR-494brain | UGAAACAUACACGGGAAACCUCUU |
| miR-496 | 1163 | 1170 | 1A,m8 | hsa-miR-496 | AUUACAUGGCCAAUCUC |
| hsa-miR-496 | AUUACAUGGCCAAUCUC | ||||
| miR-544 | 584 | 590 | 1A | hsa-miR-544 | AUUCUGCAUUUUUAGCAAGU |
| miR-93.hd/291-3p/294/295/302/372/373/520 | 619 | 625 | m8 | hsa-miR-93brain | AAAGUGCUGUUCGUGCAGGUAG |
| hsa-miR-302a | UAAGUGCUUCCAUGUUUUGGUGA | ||||
| hsa-miR-302b | UAAGUGCUUCCAUGUUUUAGUAG | ||||
| hsa-miR-302c | UAAGUGCUUCCAUGUUUCAGUGG | ||||
| hsa-miR-302d | UAAGUGCUUCCAUGUUUGAGUGU | ||||
| hsa-miR-372 | AAAGUGCUGCGACAUUUGAGCGU | ||||
| hsa-miR-373 | GAAGUGCUUCGAUUUUGGGGUGU | ||||
| hsa-miR-520e | AAAGUGCUUCCUUUUUGAGGG | ||||
| hsa-miR-520a | AAAGUGCUUCCCUUUGGACUGU | ||||
| hsa-miR-520b | AAAGUGCUUCCUUUUAGAGGG | ||||
| hsa-miR-520c | AAAGUGCUUCCUUUUAGAGGGUU | ||||
| hsa-miR-520d | AAAGUGCUUCUCUUUGGUGGGUU | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.