Gene Page: NOTCH3
Summary ?
| GeneID | 4854 |
| Symbol | NOTCH3 |
| Synonyms | CADASIL|CADASIL1|CASIL|IMF2|LMNS |
| Description | notch 3 |
| Reference | MIM:600276|HGNC:HGNC:7883|Ensembl:ENSG00000074181|HPRD:02607|Vega:OTTHUMG00000183217 |
| Gene type | protein-coding |
| Map location | 19p13.12 |
| Pascal p-value | 0.106 |
| Fetal beta | -0.018 |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 3 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs9847958 | chr3 | 23361727 | NOTCH3 | 4854 | 0.13 | trans |
Section II. Transcriptome annotation
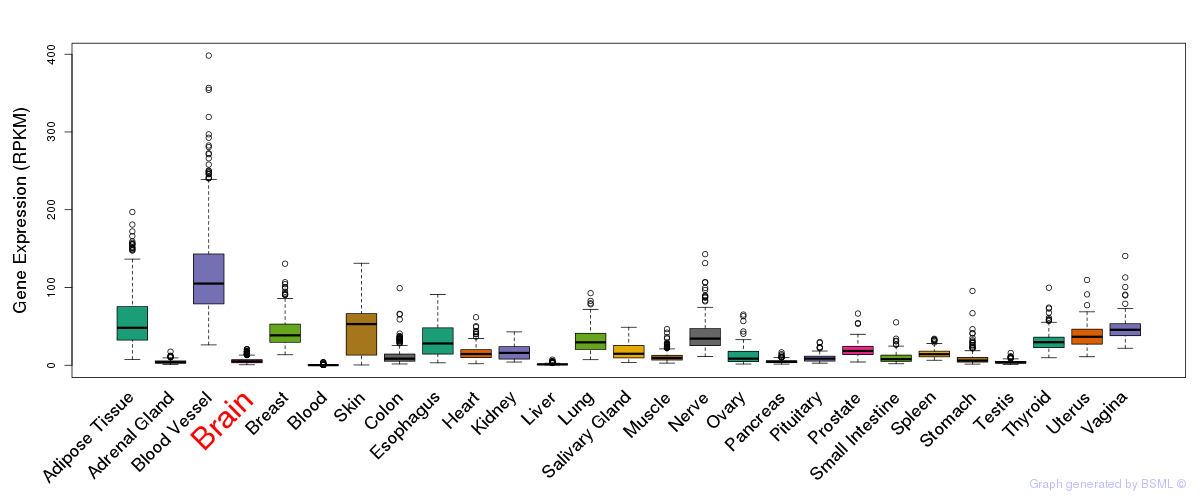
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| PTPRN | 0.90 | 0.89 |
| GALNT9 | 0.86 | 0.87 |
| SYP | 0.85 | 0.89 |
| DUSP3 | 0.85 | 0.86 |
| RASGRF1 | 0.85 | 0.86 |
| TRPM2 | 0.85 | 0.87 |
| SYNGR1 | 0.85 | 0.83 |
| GRIN1 | 0.85 | 0.83 |
| CNTNAP1 | 0.85 | 0.86 |
| MFSD4 | 0.85 | 0.85 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| RBMX2 | -0.43 | -0.52 |
| KIAA1949 | -0.43 | -0.30 |
| SH3BP2 | -0.43 | -0.45 |
| C21orf57 | -0.42 | -0.42 |
| FAM36A | -0.42 | -0.36 |
| TUBB2B | -0.42 | -0.42 |
| SH2D2A | -0.42 | -0.46 |
| GTF3C6 | -0.41 | -0.39 |
| TRAF4 | -0.41 | -0.44 |
| FADS2 | -0.41 | -0.34 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004872 | receptor activity | IEA | - | |
| GO:0005509 | calcium ion binding | IEA | - | |
| GO:0005515 | protein binding | IPI | 17292860 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0048663 | neuron fate commitment | IEA | neuron (GO term level: 9) | - |
| GO:0030900 | forebrain development | IEA | Brain (GO term level: 8) | - |
| GO:0045665 | negative regulation of neuron differentiation | IEA | neuron (GO term level: 10) | - |
| GO:0006355 | regulation of transcription, DNA-dependent | IEA | - | |
| GO:0006350 | transcription | IEA | - | |
| GO:0007219 | Notch signaling pathway | IEA | - | |
| GO:0009653 | anatomical structure morphogenesis | TAS | 7698746 | |
| GO:0007275 | multicellular organismal development | IEA | - | |
| GO:0050793 | regulation of developmental process | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | IEA | - | |
| GO:0016021 | integral to membrane | IEA | - | |
| GO:0005886 | plasma membrane | IEA | - | |
| GO:0005887 | integral to plasma membrane | TAS | 7835890 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG DORSO VENTRAL AXIS FORMATION | 25 | 21 | All SZGR 2.0 genes in this pathway |
| KEGG NOTCH SIGNALING PATHWAY | 47 | 35 | All SZGR 2.0 genes in this pathway |
| PID NOTCH PATHWAY | 59 | 49 | All SZGR 2.0 genes in this pathway |
| REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | 29 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME PRE NOTCH EXPRESSION AND PROCESSING | 44 | 24 | All SZGR 2.0 genes in this pathway |
| REACTOME PRE NOTCH PROCESSING IN GOLGI | 16 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY NOTCH3 | 12 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME GENERIC TRANSCRIPTION PATHWAY | 352 | 181 | All SZGR 2.0 genes in this pathway |
| REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | 13 | 8 | All SZGR 2.0 genes in this pathway |
| REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | 12 | 8 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY NOTCH | 103 | 64 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA UP | 783 | 507 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 TARGETS DN | 260 | 143 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC3 TARGETS DN | 536 | 332 | All SZGR 2.0 genes in this pathway |
| LIU CDX2 TARGETS UP | 36 | 22 | All SZGR 2.0 genes in this pathway |
| PEREZ TP63 TARGETS | 355 | 243 | All SZGR 2.0 genes in this pathway |
| PEREZ TP53 AND TP63 TARGETS | 205 | 145 | All SZGR 2.0 genes in this pathway |
| SCHEIDEREIT IKK INTERACTING PROTEINS | 58 | 45 | All SZGR 2.0 genes in this pathway |
| KOMMAGANI TP63 GAMMA TARGETS | 9 | 7 | All SZGR 2.0 genes in this pathway |
| PATIL LIVER CANCER | 747 | 453 | All SZGR 2.0 genes in this pathway |
| JEON SMAD6 TARGETS DN | 19 | 14 | All SZGR 2.0 genes in this pathway |
| SHIN B CELL LYMPHOMA CLUSTER 7 | 27 | 22 | All SZGR 2.0 genes in this pathway |
| CERVERA SDHB TARGETS 2 | 114 | 76 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER DENA UP | 60 | 40 | All SZGR 2.0 genes in this pathway |
| DEBIASI APOPTOSIS BY REOVIRUS INFECTION DN | 287 | 208 | All SZGR 2.0 genes in this pathway |
| RAMASWAMY METASTASIS UP | 66 | 43 | All SZGR 2.0 genes in this pathway |
| SATO SILENCED BY DEACETYLATION IN PANCREATIC CANCER | 50 | 30 | All SZGR 2.0 genes in this pathway |
| MARCHINI TRABECTEDIN RESISTANCE DN | 49 | 34 | All SZGR 2.0 genes in this pathway |
| DOUGLAS BMI1 TARGETS UP | 566 | 371 | All SZGR 2.0 genes in this pathway |
| MCCOLLUM GELDANAMYCIN RESISTANCE DN | 7 | 6 | All SZGR 2.0 genes in this pathway |
| BRUECKNER TARGETS OF MIRLET7A3 UP | 111 | 69 | All SZGR 2.0 genes in this pathway |
| VART KSHV INFECTION ANGIOGENIC MARKERS DN | 138 | 92 | All SZGR 2.0 genes in this pathway |
| VILIMAS NOTCH1 TARGETS UP | 52 | 41 | All SZGR 2.0 genes in this pathway |
| MCMURRAY TP53 HRAS COOPERATION RESPONSE DN | 67 | 46 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K27ME3 | 269 | 159 | All SZGR 2.0 genes in this pathway |
| CROONQUIST STROMAL STIMULATION UP | 60 | 42 | All SZGR 2.0 genes in this pathway |
| CROONQUIST NRAS VS STROMAL STIMULATION DN | 99 | 65 | All SZGR 2.0 genes in this pathway |
| JISON SICKLE CELL DISEASE UP | 181 | 106 | All SZGR 2.0 genes in this pathway |
| CHIARETTI T ALL REFRACTORY TO THERAPY | 30 | 18 | All SZGR 2.0 genes in this pathway |
| LI INDUCED T TO NATURAL KILLER DN | 116 | 83 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE UP | 857 | 456 | All SZGR 2.0 genes in this pathway |
| CHYLA CBFA2T3 TARGETS UP | 387 | 225 | All SZGR 2.0 genes in this pathway |
| FUKUSHIMA TNFSF11 TARGETS | 16 | 14 | All SZGR 2.0 genes in this pathway |
| WANG TNF TARGETS | 24 | 17 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS UP | 745 | 475 | All SZGR 2.0 genes in this pathway |
| KATSANOU ELAVL1 TARGETS UP | 169 | 105 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-1/206 | 658 | 664 | 1A | hsa-miR-1 | UGGAAUGUAAAGAAGUAUGUA |
| hsa-miR-206SZ | UGGAAUGUAAGGAAGUGUGUGG | ||||
| hsa-miR-613 | AGGAAUGUUCCUUCUUUGCC | ||||
| miR-150 | 618 | 624 | m8 | hsa-miR-150 | UCUCCCAACCCUUGUACCAGUG |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.