Gene Page: OPRK1
Summary ?
| GeneID | 4986 |
| Symbol | OPRK1 |
| Synonyms | K-OR-1|KOR|KOR-1|OPRK |
| Description | opioid receptor, kappa 1 |
| Reference | MIM:165196|HGNC:HGNC:8154|Ensembl:ENSG00000082556|HPRD:01309|Vega:OTTHUMG00000164276 |
| Gene type | protein-coding |
| Map location | 8q11.2 |
| Pascal p-value | 0.939 |
| DMG | 1 (# studies) |
| eGene | Cerebellar Hemisphere Meta |
| Support | GPCR SIGNALLING |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg09999109 | 8 | 54164310 | OPRK1 | 4.82E-7 | -0.392 | 0.006 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
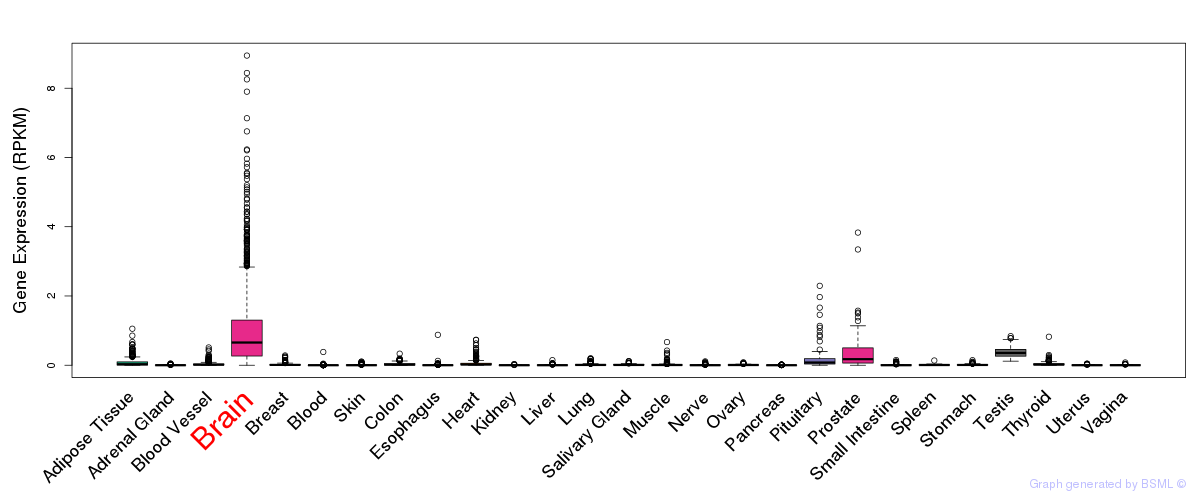
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| YIPF3 | 0.91 | 0.88 |
| PSMF1 | 0.89 | 0.86 |
| PSMD3 | 0.89 | 0.88 |
| DDOST | 0.89 | 0.89 |
| ERAL1 | 0.89 | 0.85 |
| EIF2B2 | 0.89 | 0.84 |
| TRAP1 | 0.88 | 0.84 |
| EIF3I | 0.88 | 0.86 |
| PSMC4 | 0.88 | 0.85 |
| DCTN2 | 0.88 | 0.85 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MT-CO2 | -0.78 | -0.77 |
| AF347015.31 | -0.77 | -0.77 |
| AF347015.8 | -0.76 | -0.77 |
| MT-CYB | -0.75 | -0.77 |
| AF347015.33 | -0.74 | -0.76 |
| AF347015.21 | -0.74 | -0.78 |
| AF347015.2 | -0.74 | -0.77 |
| AF347015.27 | -0.73 | -0.77 |
| AF347015.26 | -0.71 | -0.76 |
| AF347015.15 | -0.69 | -0.73 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004872 | receptor activity | IEA | - | |
| GO:0005515 | protein binding | IPI | 16431922 | |
| GO:0004987 | kappa-opioid receptor activity | IEA | - | |
| GO:0004987 | kappa-opioid receptor activity | TAS | 8755601 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007268 | synaptic transmission | TAS | neuron, Synap, Neurotransmitter (GO term level: 6) | 7624359 |
| GO:0007186 | G-protein coupled receptor protein signaling pathway | IEA | - | |
| GO:0007193 | G-protein signaling, adenylate cyclase inhibiting pathway | TAS | 7624359 | |
| GO:0007165 | signal transduction | IEA | - | |
| GO:0007610 | behavior | TAS | 7624359 | |
| GO:0007600 | sensory perception | TAS | 7624359 | |
| GO:0006955 | immune response | TAS | 8755601 | |
| GO:0019079 | viral genome replication | TAS | 8755601 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0016021 | integral to membrane | IEA | - | |
| GO:0005886 | plasma membrane | IEA | - | |
| GO:0005887 | integral to plasma membrane | TAS | 7624359 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG NEUROACTIVE LIGAND RECEPTOR INTERACTION | 272 | 195 | All SZGR 2.0 genes in this pathway |
| BIOCARTA DREAM PATHWAY | 14 | 13 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY GPCR | 920 | 449 | All SZGR 2.0 genes in this pathway |
| REACTOME PEPTIDE LIGAND BINDING RECEPTORS | 188 | 108 | All SZGR 2.0 genes in this pathway |
| REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | 305 | 177 | All SZGR 2.0 genes in this pathway |
| REACTOME GPCR DOWNSTREAM SIGNALING | 805 | 368 | All SZGR 2.0 genes in this pathway |
| REACTOME G ALPHA I SIGNALLING EVENTS | 195 | 114 | All SZGR 2.0 genes in this pathway |
| REACTOME GPCR LIGAND BINDING | 408 | 246 | All SZGR 2.0 genes in this pathway |
| ODONNELL METASTASIS UP | 82 | 58 | All SZGR 2.0 genes in this pathway |
| BENPORATH SUZ12 TARGETS | 1038 | 678 | All SZGR 2.0 genes in this pathway |
| KAUFFMANN DNA REPLICATION GENES | 147 | 87 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 48HR DN | 504 | 323 | All SZGR 2.0 genes in this pathway |
| SATO SILENCED BY METHYLATION IN PANCREATIC CANCER 1 | 419 | 273 | All SZGR 2.0 genes in this pathway |
| IWANAGA CARCINOGENESIS BY KRAS PTEN DN | 353 | 226 | All SZGR 2.0 genes in this pathway |
| LABBE TGFB1 TARGETS UP | 102 | 64 | All SZGR 2.0 genes in this pathway |
| LABBE TARGETS OF TGFB1 AND WNT3A UP | 111 | 70 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| ROME INSULIN TARGETS IN MUSCLE DN | 204 | 114 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN IPS LCP WITH H3K4ME3 | 174 | 100 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN ES LCP WITH H3K4ME3 | 142 | 80 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 30MIN DN | 150 | 99 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-381 | 3517 | 3523 | 1A | hsa-miR-381 | UAUACAAGGGCAAGCUCUCUGU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.