Gene Page: CCDC53
Summary ?
| GeneID | 51019 |
| Symbol | CCDC53 |
| Synonyms | CGI-116 |
| Description | coiled-coil domain containing 53 |
| Reference | HGNC:HGNC:24256|Ensembl:ENSG00000120860|HPRD:13031|Vega:OTTHUMG00000168187 |
| Gene type | protein-coding |
| Map location | 12q23.2 |
| Pascal p-value | 0.606 |
| Sherlock p-value | 0.059 |
| Fetal beta | -0.588 |
| DMG | 1 (# studies) |
| eGene | Cortex Hypothalamus Nucleus accumbens basal ganglia Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0105 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg16884940 | 12 | 102455204 | CCDC53 | 3.59E-8 | -0.021 | 1.05E-5 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
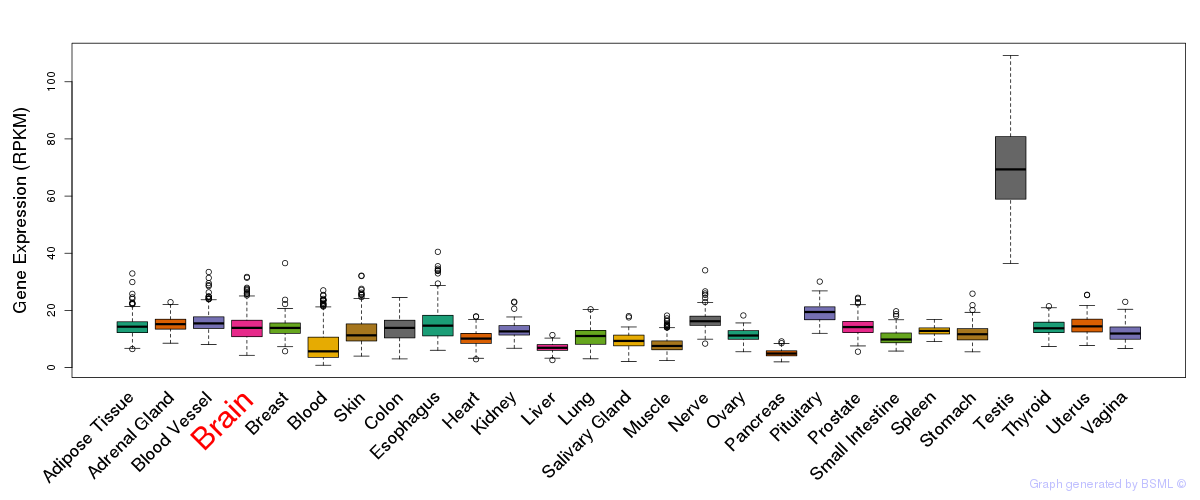
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003674 | molecular_function | ND | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0008150 | biological_process | ND | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005575 | cellular_component | ND | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ABI2 | ABI-2 | ABI2B | AIP-1 | AblBP3 | SSH3BP2 | argBPIA | argBPIB | abl interactor 2 | Two-hybrid | BioGRID | 16189514 |
| GDF9 | - | growth differentiation factor 9 | Two-hybrid | BioGRID | 16169070 |
| HSBP1 | DKFZp686D1664 | DKFZp686O24200 | NPC-A-13 | heat shock factor binding protein 1 | Two-hybrid | BioGRID | 16189514 |
| KRT6A | CK6A | CK6C | CK6D | K6A | K6C | K6D | KRT6C | KRT6D | keratin 6A | Two-hybrid | BioGRID | 16189514 |
| LAMC1 | LAMB2 | MGC87297 | laminin, gamma 1 (formerly LAMB2) | Two-hybrid | BioGRID | 16169070 |
| LUC7L2 | CGI-59 | CGI-74 | FLJ10657 | LUC7B2 | LUC7-like 2 (S. cerevisiae) | Two-hybrid | BioGRID | 16169070 |
| MCRS1 | ICP22BP | INO80Q | MCRS2 | MSP58 | P78 | microspherule protein 1 | Two-hybrid | BioGRID | 16189514 |
| NDC80 | HEC | HEC1 | KNTC2 | TID3 | hsNDC80 | NDC80 homolog, kinetochore complex component (S. cerevisiae) | Two-hybrid | BioGRID | 16189514 |
| NUP62 | DKFZp547L134 | FLJ20822 | FLJ43869 | IBSN | MGC841 | SNDI | p62 | nucleoporin 62kDa | Two-hybrid | BioGRID | 16189514 |
| OPTN | FIP2 | GLC1E | HIP7 | HYPL | NRP | TFIIIA-INTP | optineurin | Two-hybrid | BioGRID | 16189514 |
| PBXIP1 | HPIP | pre-B-cell leukemia homeobox interacting protein 1 | Two-hybrid | BioGRID | 16189514 |
| PCM1 | PTC4 | pericentriolar material 1 | Two-hybrid | BioGRID | 16189514 |
| PLDN | PA | PALLID | pallidin homolog (mouse) | Two-hybrid | BioGRID | 16189514 |
| PTN | HARP | HBGF8 | HBNF | NEGF1 | pleiotrophin | Two-hybrid | BioGRID | 16169070 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| DAVICIONI MOLECULAR ARMS VS ERMS UP | 332 | 228 | All SZGR 2.0 genes in this pathway |
| LIU CMYB TARGETS UP | 165 | 106 | All SZGR 2.0 genes in this pathway |
| LIU VMYB TARGETS UP | 127 | 78 | All SZGR 2.0 genes in this pathway |
| UDAYAKUMAR MED1 TARGETS DN | 240 | 171 | All SZGR 2.0 genes in this pathway |
| HAHTOLA MYCOSIS FUNGOIDES CD4 DN | 116 | 71 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA DN | 394 | 258 | All SZGR 2.0 genes in this pathway |
| TERAMOTO OPN TARGETS CLUSTER 5 | 7 | 5 | All SZGR 2.0 genes in this pathway |
| KIM MYCN AMPLIFICATION TARGETS DN | 103 | 59 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS UP | 769 | 437 | All SZGR 2.0 genes in this pathway |
| SWEET KRAS TARGETS UP | 84 | 51 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| BOYAULT LIVER CANCER SUBCLASS G6 UP | 65 | 43 | All SZGR 2.0 genes in this pathway |
| CHIANG LIVER CANCER SUBCLASS CTNNB1 UP | 176 | 110 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA CLASSES DN | 210 | 141 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN ES ICP WITH H3K4ME3 | 718 | 401 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN NPC ICP WITH H3K4ME3 | 445 | 257 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS GROWING | 243 | 155 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS CONFLUENT | 567 | 365 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE LATE | 1137 | 655 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 1ST EGF PULSE ONLY | 1839 | 928 | All SZGR 2.0 genes in this pathway |