Gene Page: LRRC1
Summary ?
| GeneID | 55227 |
| Symbol | LRRC1 |
| Synonyms | LANO|dJ523E19.1 |
| Description | leucine rich repeat containing 1 |
| Reference | MIM:608195|HGNC:HGNC:14307|Ensembl:ENSG00000137269|Vega:OTTHUMG00000014885 |
| Gene type | protein-coding |
| Map location | 6p12.1 |
| Pascal p-value | 0.115 |
| Sherlock p-value | 0.112 |
| DEG p-value | DEG:Zhao_2015:p=6.97e-04:q=0.0984 |
| Fetal beta | 1.348 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DEG:Zhao_2015 | RNA Sequencing analysis | Transcriptome sequencing and genome-wide association analyses reveal lysosomal function and actin cytoskeleton remodeling in schizophrenia and bipolar disorder. | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
| GSMA_IIE | Genome scan meta-analysis (European-ancestry samples) | Psr: 0.04433 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg15286372 | 6 | 53659348 | LRRC1 | 4.47E-8 | -0.015 | 1.22E-5 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs1346726 | chr8 | 27335218 | LRRC1 | 55227 | 0.2 | trans |
Section II. Transcriptome annotation
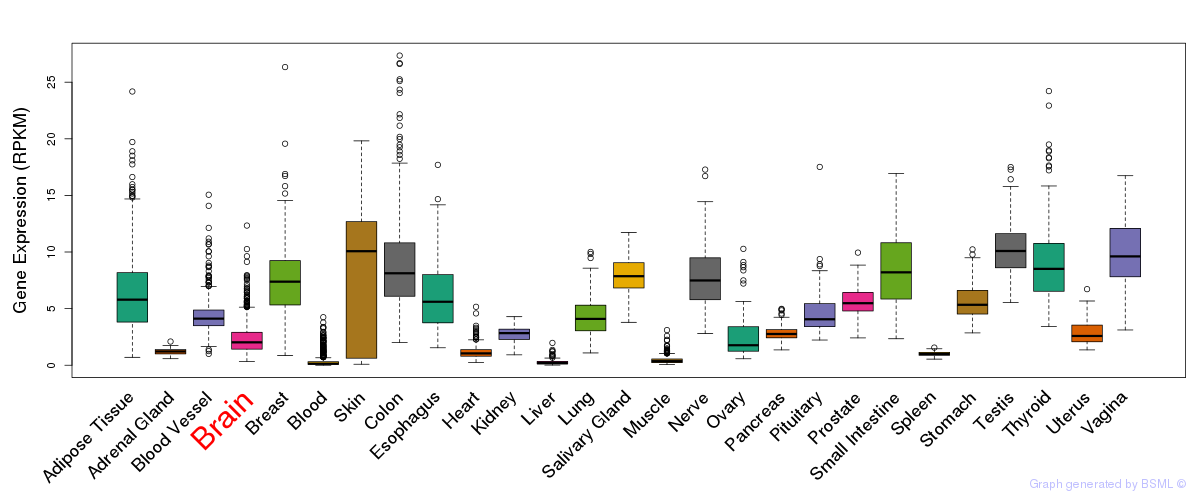
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| RBM14 | 0.93 | 0.90 |
| CCDC71 | 0.93 | 0.94 |
| PCIF1 | 0.93 | 0.92 |
| ZNF574 | 0.92 | 0.89 |
| USP21 | 0.92 | 0.91 |
| TOR2A | 0.92 | 0.91 |
| SMPD4 | 0.91 | 0.89 |
| DCAKD | 0.91 | 0.89 |
| FAM127C | 0.91 | 0.88 |
| RSAD1 | 0.91 | 0.89 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.27 | -0.69 | -0.82 |
| AF347015.31 | -0.69 | -0.81 |
| MT-CO2 | -0.67 | -0.80 |
| AF347015.8 | -0.66 | -0.81 |
| MT-CYB | -0.66 | -0.80 |
| AF347015.33 | -0.65 | -0.77 |
| C5orf53 | -0.64 | -0.69 |
| AF347015.15 | -0.63 | -0.79 |
| AF347015.21 | -0.63 | -0.85 |
| AF347015.2 | -0.60 | -0.77 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005515 | protein binding | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0016020 | membrane | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| ONKEN UVEAL MELANOMA DN | 526 | 357 | All SZGR 2.0 genes in this pathway |
| HORIUCHI WTAP TARGETS UP | 306 | 188 | All SZGR 2.0 genes in this pathway |
| OSWALD HEMATOPOIETIC STEM CELL IN COLLAGEN GEL DN | 275 | 168 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 AND HDAC2 TARGETS DN | 232 | 139 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE FIMA DN | 284 | 156 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 2 DN | 464 | 276 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL AND PROGENITOR | 681 | 420 | All SZGR 2.0 genes in this pathway |
| MASSARWEH TAMOXIFEN RESISTANCE UP | 578 | 341 | All SZGR 2.0 genes in this pathway |
| CHIANG LIVER CANCER SUBCLASS PROLIFERATION UP | 178 | 108 | All SZGR 2.0 genes in this pathway |
| LOPEZ EPITHELIOID MESOTHELIOMA | 17 | 11 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA UP | 207 | 143 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA CLASSES UP | 605 | 377 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 24HR DN | 505 | 328 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 1ST EGF PULSE ONLY | 1839 | 928 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-124.1 | 595 | 601 | m8 | hsa-miR-124a | UUAAGGCACGCGGUGAAUGCCA |
| miR-124/506 | 594 | 601 | 1A,m8 | hsa-miR-506 | UAAGGCACCCUUCUGAGUAGA |
| hsa-miR-124brain | UAAGGCACGCGGUGAAUGCC | ||||
| miR-134 | 1255 | 1261 | m8 | hsa-miR-134brain | UGUGACUGGUUGACCAGAGGG |
| miR-142-3p | 245 | 251 | m8 | hsa-miR-142-3p | UGUAGUGUUUCCUACUUUAUGGA |
| miR-22 | 434 | 440 | m8 | hsa-miR-22brain | AAGCUGCCAGUUGAAGAACUGU |
| miR-381 | 464 | 470 | 1A | hsa-miR-381 | UAUACAAGGGCAAGCUCUCUGU |
| miR-495 | 502 | 508 | 1A | hsa-miR-495brain | AAACAAACAUGGUGCACUUCUUU |
| miR-544 | 919 | 925 | 1A | hsa-miR-544 | AUUCUGCAUUUUUAGCAAGU |
| miR-9 | 794 | 800 | m8 | hsa-miR-9SZ | UCUUUGGUUAUCUAGCUGUAUGA |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.