Gene Page: PSMD2
Summary ?
| GeneID | 5708 |
| Symbol | PSMD2 |
| Synonyms | P97|RPN1|S2|TRAP2 |
| Description | proteasome 26S subunit, non-ATPase 2 |
| Reference | MIM:606223|HGNC:HGNC:9559|Ensembl:ENSG00000175166|HPRD:05870|Vega:OTTHUMG00000156796 |
| Gene type | protein-coding |
| Map location | 3q27.1 |
| Pascal p-value | 0.105 |
| Sherlock p-value | 0.003 |
| TADA p-value | 0.014 |
| Fetal beta | -0.404 |
| eGene | Cerebellar Hemisphere Nucleus accumbens basal ganglia Myers' cis & trans |
| Support | G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.humanPSD G2Cdb.humanPSP CompositeSet Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DNM:Fromer_2014 | Whole Exome Sequencing analysis | This study reported a WES study of 623 schizophrenia trios, reporting DNMs using genomic DNA. | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0069 |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| PSMD2 | chr3 | 184025295 | A | G | NM_002808 | p.729M>V | missense | Schizophrenia | DNM:Fromer_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs17432613 | chr1 | 18346091 | PSMD2 | 5708 | 0.19 | trans |
Section II. Transcriptome annotation
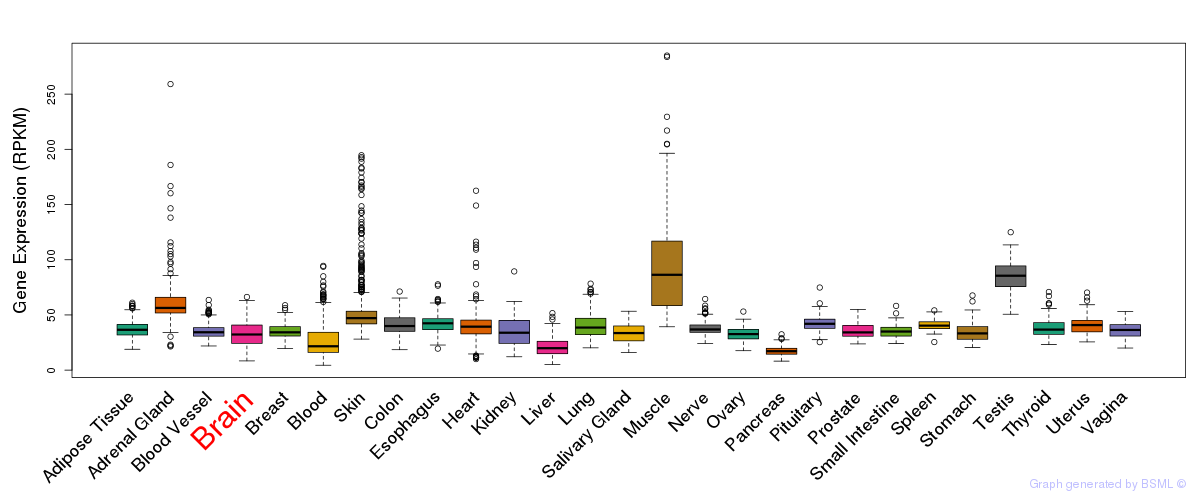
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| FGD1 | 0.91 | 0.92 |
| SARM1 | 0.89 | 0.91 |
| GLT25D1 | 0.89 | 0.91 |
| DTX4 | 0.89 | 0.92 |
| ASXL1 | 0.89 | 0.90 |
| UBAP2 | 0.89 | 0.89 |
| CCDC120 | 0.89 | 0.90 |
| FRMD4A | 0.88 | 0.87 |
| GPR173 | 0.88 | 0.90 |
| ZMYM3 | 0.88 | 0.88 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.31 | -0.74 | -0.87 |
| AF347015.27 | -0.73 | -0.86 |
| S100B | -0.73 | -0.83 |
| MT-CO2 | -0.73 | -0.87 |
| AIFM3 | -0.72 | -0.75 |
| AF347015.33 | -0.72 | -0.85 |
| TSC22D4 | -0.71 | -0.77 |
| HEPN1 | -0.71 | -0.75 |
| MT-CYB | -0.71 | -0.85 |
| C5orf53 | -0.70 | -0.74 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005488 | binding | IEA | - | |
| GO:0005515 | protein binding | IPI | 11278995 | |
| GO:0030234 | enzyme regulator activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0042176 | regulation of protein catabolic process | IEA | - | |
| GO:0031145 | anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | EXP | 11285280 | |
| GO:0051436 | negative regulation of ubiquitin-protein ligase activity during mitotic cell cycle | EXP | 15029244 | |
| GO:0051437 | positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | EXP | 12791267 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000502 | proteasome complex | TAS | 8811196 | |
| GO:0005829 | cytosol | IEA | - | |
| GO:0005838 | proteasome regulatory particle | TAS | 8774743 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| C1orf103 | FLJ11269 | RIF1 | RP11-96K19.1 | chromosome 1 open reading frame 103 | Two-hybrid | BioGRID | 16169070 |
| MCC | DKFZp762O1615 | FLJ38893 | FLJ46755 | MCC1 | mutated in colorectal cancers | Affinity Capture-MS | BioGRID | 17353931 |
| NEK6 | SID6-1512 | NIMA (never in mitosis gene a)-related kinase 6 | Affinity Capture-MS | BioGRID | 17353931 |
| PAAF1 | FLJ11848 | PAAF | Rpn14 | WDR71 | proteasomal ATPase-associated factor 1 | Affinity Capture-MS | BioGRID | 17353931 |
| PSMC1 | MGC24583 | MGC8541 | P26S4 | S4 | p56 | proteasome (prosome, macropain) 26S subunit, ATPase, 1 | - | HPRD,BioGRID | 10625621 |
| PSMC2 | MGC3004 | MSS1 | Nbla10058 | S7 | proteasome (prosome, macropain) 26S subunit, ATPase, 2 | - | HPRD,BioGRID | 10625621 |
| PSMC4 | MGC13687 | MGC23214 | MGC8570 | MIP224 | S6 | TBP7 | proteasome (prosome, macropain) 26S subunit, ATPase, 4 | Affinity Capture-MS | BioGRID | 17353931 |
| PSMD10 | dJ889N15.2 | p28 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 10 | Affinity Capture-MS | BioGRID | 17353931 |
| PSMD13 | HSPC027 | Rpn9 | S11 | p40.5 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 13 | Affinity Capture-MS | BioGRID | 17353931 |
| PSMD5 | KIAA0072 | MGC23145 | S5B | proteasome (prosome, macropain) 26S subunit, non-ATPase, 5 | - | HPRD,BioGRID | 10625621 |
| PSMD6 | KIAA0107 | Rpn7 | S10 | SGA-113M | p44S10 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 6 | Affinity Capture-MS | BioGRID | 17353931 |
| PTN | HARP | HBGF8 | HBNF | NEGF1 | pleiotrophin | Two-hybrid | BioGRID | 16169070 |
| TNFRSF1A | CD120a | FPF | MGC19588 | TBP1 | TNF-R | TNF-R-I | TNF-R55 | TNFAR | TNFR1 | TNFR55 | TNFR60 | p55 | p55-R | p60 | tumor necrosis factor receptor superfamily, member 1A | - | HPRD,BioGRID | 7601280 |9126987 |
| UBE3C | KIAA0010 | KIAA10 | ubiquitin protein ligase E3C | - | HPRD,BioGRID | 12692129 |
| UBLCP1 | CPUB1 | FLJ25267 | MGC10067 | ubiquitin-like domain containing CTD phosphatase 1 | Two-hybrid | BioGRID | 16189514 |
| UNC119 | HRG4 | unc-119 homolog (C. elegans) | Two-hybrid | BioGRID | 16169070 |
| WDR8 | FLJ20430 | MGC99569 | WD repeat domain 8 | Affinity Capture-MS | BioGRID | 17353931 |
| ZBTB16 | PLZF | ZNF145 | zinc finger and BTB domain containing 16 | Two-hybrid | BioGRID | 16169070 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG PROTEASOME | 48 | 28 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY WNT | 65 | 41 | All SZGR 2.0 genes in this pathway |
| REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | 48 | 29 | All SZGR 2.0 genes in this pathway |
| REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | 76 | 48 | All SZGR 2.0 genes in this pathway |
| REACTOME ER PHAGOSOME PATHWAY | 61 | 40 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE | 421 | 253 | All SZGR 2.0 genes in this pathway |
| REACTOME ORC1 REMOVAL FROM CHROMATIN | 67 | 37 | All SZGR 2.0 genes in this pathway |
| REACTOME DOWNSTREAM SIGNALING EVENTS OF B CELL RECEPTOR BCR | 97 | 66 | All SZGR 2.0 genes in this pathway |
| REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | 64 | 43 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY THE B CELL RECEPTOR BCR | 126 | 90 | All SZGR 2.0 genes in this pathway |
| REACTOME P53 INDEPENDENT G1 S DNA DAMAGE CHECKPOINT | 51 | 31 | All SZGR 2.0 genes in this pathway |
| REACTOME CDK MEDIATED PHOSPHORYLATION AND REMOVAL OF CDC6 | 48 | 27 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | 200 | 136 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | 49 | 33 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF APOPTOSIS | 58 | 34 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE MITOTIC | 325 | 185 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE CHECKPOINTS | 124 | 70 | All SZGR 2.0 genes in this pathway |
| REACTOME CYCLIN E ASSOCIATED EVENTS DURING G1 S TRANSITION | 65 | 40 | All SZGR 2.0 genes in this pathway |
| REACTOME P53 DEPENDENT G1 DNA DAMAGE RESPONSE | 57 | 35 | All SZGR 2.0 genes in this pathway |
| REACTOME M G1 TRANSITION | 81 | 45 | All SZGR 2.0 genes in this pathway |
| REACTOME G1 S TRANSITION | 112 | 63 | All SZGR 2.0 genes in this pathway |
| REACTOME CDT1 ASSOCIATION WITH THE CDC6 ORC ORIGIN COMPLEX | 56 | 31 | All SZGR 2.0 genes in this pathway |
| REACTOME SYNTHESIS OF DNA | 92 | 53 | All SZGR 2.0 genes in this pathway |
| REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | 51 | 30 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF MRNA | 284 | 128 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF RNA | 330 | 155 | All SZGR 2.0 genes in this pathway |
| REACTOME MITOTIC G1 G1 S PHASES | 137 | 79 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF MITOTIC CELL CYCLE | 85 | 46 | All SZGR 2.0 genes in this pathway |
| REACTOME MITOTIC M M G1 PHASES | 172 | 98 | All SZGR 2.0 genes in this pathway |
| REACTOME ASSEMBLY OF THE PRE REPLICATIVE COMPLEX | 65 | 36 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | 84 | 50 | All SZGR 2.0 genes in this pathway |
| REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | 53 | 31 | All SZGR 2.0 genes in this pathway |
| REACTOME DNA REPLICATION | 192 | 110 | All SZGR 2.0 genes in this pathway |
| REACTOME APOPTOSIS | 148 | 94 | All SZGR 2.0 genes in this pathway |
| REACTOME HIV INFECTION | 207 | 122 | All SZGR 2.0 genes in this pathway |
| REACTOME HOST INTERACTIONS OF HIV FACTORS | 132 | 81 | All SZGR 2.0 genes in this pathway |
| REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | 72 | 38 | All SZGR 2.0 genes in this pathway |
| REACTOME APC C CDC20 MEDIATED DEGRADATION OF MITOTIC PROTEINS | 73 | 40 | All SZGR 2.0 genes in this pathway |
| REACTOME AUTODEGRADATION OF CDH1 BY CDH1 APC C | 64 | 34 | All SZGR 2.0 genes in this pathway |
| REACTOME SCF BETA TRCP MEDIATED DEGRADATION OF EMI1 | 51 | 32 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME ADAPTIVE IMMUNE SYSTEM | 539 | 350 | All SZGR 2.0 genes in this pathway |
| REACTOME CLASS I MHC MEDIATED ANTIGEN PROCESSING PRESENTATION | 251 | 156 | All SZGR 2.0 genes in this pathway |
| REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | 212 | 129 | All SZGR 2.0 genes in this pathway |
| REACTOME S PHASE | 109 | 66 | All SZGR 2.0 genes in this pathway |
| REACTOME SCFSKP2 MEDIATED DEGRADATION OF P27 P21 | 56 | 34 | All SZGR 2.0 genes in this pathway |
| REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | 52 | 30 | All SZGR 2.0 genes in this pathway |
| WINTER HYPOXIA UP | 92 | 57 | All SZGR 2.0 genes in this pathway |
| NAKAMURA TUMOR ZONE PERIPHERAL VS CENTRAL UP | 285 | 181 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| BORCZUK MALIGNANT MESOTHELIOMA UP | 305 | 185 | All SZGR 2.0 genes in this pathway |
| BASAKI YBX1 TARGETS UP | 290 | 177 | All SZGR 2.0 genes in this pathway |
| TIEN INTESTINE PROBIOTICS 24HR UP | 557 | 331 | All SZGR 2.0 genes in this pathway |
| NADERI BREAST CANCER PROGNOSIS UP | 50 | 26 | All SZGR 2.0 genes in this pathway |
| TOMIDA METASTASIS UP | 26 | 13 | All SZGR 2.0 genes in this pathway |
| DARWICHE PAPILLOMA RISK HIGH VS LOW DN | 32 | 21 | All SZGR 2.0 genes in this pathway |
| WEI MYCN TARGETS WITH E BOX | 795 | 478 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 6HR DN | 514 | 330 | All SZGR 2.0 genes in this pathway |
| HE PTEN TARGETS DN | 7 | 6 | All SZGR 2.0 genes in this pathway |
| BASSO B LYMPHOCYTE NETWORK | 143 | 96 | All SZGR 2.0 genes in this pathway |
| PENG LEUCINE DEPRIVATION DN | 187 | 122 | All SZGR 2.0 genes in this pathway |
| VANTVEER BREAST CANCER METASTASIS DN | 121 | 65 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| RHODES UNDIFFERENTIATED CANCER | 69 | 44 | All SZGR 2.0 genes in this pathway |
| ZHU CMV ALL UP | 120 | 89 | All SZGR 2.0 genes in this pathway |
| ZHU CMV 24 HR UP | 93 | 65 | All SZGR 2.0 genes in this pathway |
| PELLICCIOTTA HDAC IN ANTIGEN PRESENTATION UP | 64 | 40 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER UP | 973 | 570 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER TUMOR VS NORMAL ADJACENT TISSUE UP | 863 | 514 | All SZGR 2.0 genes in this pathway |
| MITSIADES RESPONSE TO APLIDIN DN | 249 | 165 | All SZGR 2.0 genes in this pathway |
| ITO PTTG1 TARGETS DN | 9 | 7 | All SZGR 2.0 genes in this pathway |
| CHUNG BLISTER CYTOTOXICITY UP | 134 | 84 | All SZGR 2.0 genes in this pathway |
| CHANG CORE SERUM RESPONSE UP | 212 | 128 | All SZGR 2.0 genes in this pathway |
| HSIAO HOUSEKEEPING GENES | 389 | 245 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER SUBCLASS S1 | 237 | 159 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS OLIGODENDROCYTE NUMBER CORR UP | 756 | 494 | All SZGR 2.0 genes in this pathway |
| KIM BIPOLAR DISORDER OLIGODENDROCYTE DENSITY CORR UP | 682 | 440 | All SZGR 2.0 genes in this pathway |
| FIGUEROA AML METHYLATION CLUSTER 6 DN | 38 | 19 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-495 | 112 | 118 | 1A | hsa-miR-495brain | AAACAAACAUGGUGCACUUCUUU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.