Gene Page: ACTB
Summary ?
| GeneID | 60 |
| Symbol | ACTB |
| Synonyms | BRWS1|PS1TP5BP1 |
| Description | actin, beta |
| Reference | MIM:102630|HGNC:HGNC:132|Ensembl:ENSG00000075624|HPRD:00032|Vega:OTTHUMG00000023268 |
| Gene type | protein-coding |
| Map location | 7p22 |
| Pascal p-value | 0.005 |
| Sherlock p-value | 0.495 |
| Fetal beta | 1.4 |
| eGene | Myers' cis & trans |
| Support | STRUCTURAL PLASTICITY CompositeSet Darnell FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenics,schizophrenias | Click to show details |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0551 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs10409262 | chr19 | 58154954 | ACTB | 60 | 0.04 | trans |
Section II. Transcriptome annotation
General gene expression (GTEx)
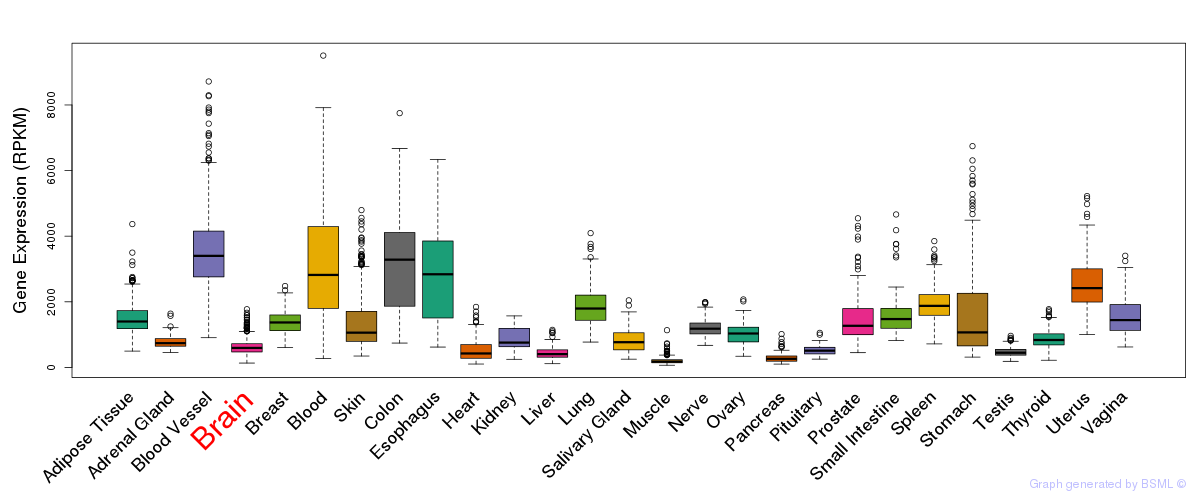
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0005515 | protein binding | IEA | - | |
| GO:0005515 | protein binding | IPI | 17342765 | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0005198 | structural molecule activity | IEA | - | |
| GO:0005200 | structural constituent of cytoskeleton | TAS | 6202424 | |
| GO:0050998 | nitric-oxide synthase binding | IPI | 17502619 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007605 | sensory perception of sound | IEA | - | |
| GO:0006928 | cell motion | TAS | 6202424 |16130169 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005829 | cytosol | IEA | - | |
| GO:0005856 | cytoskeleton | IEA | - | |
| GO:0005856 | cytoskeleton | TAS | 16130169 | |
| GO:0005625 | soluble fraction | IEA | - | |
| GO:0005737 | cytoplasm | TAS | 16130169 | |
| GO:0035267 | NuA4 histone acetyltransferase complex | IDA | 10966108 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ABRA | STARS | actin-binding Rho activating protein | - | HPRD | 11983702 |
| ACTB | PS1TP5BP1 | actin, beta | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 9209005 |16189514 |
| ACTB | PS1TP5BP1 | actin, beta | Actin interacts with itself to form an F-actin. | BIND | 15361876 |
| ACTC1 | ACTC | CMD1R | CMH11 | actin, alpha, cardiac muscle 1 | - | HPRD | 8913639 |
| ACTG1 | ACT | ACTG | DFNA20 | DFNA26 | actin, gamma 1 | Two-hybrid | BioGRID | 16189514 |
| ARPC1B | ARC41 | p40-ARC | p41-ARC | actin related protein 2/3 complex, subunit 1B, 41kDa | - | HPRD | 11533442 |11721045 |
| C20orf20 | Eaf7 | FLJ10914 | MRG15BP | MRGBP | URCC4 | chromosome 20 open reading frame 20 | - | HPRD | 12963728 |
| CCT2 | 99D8.1 | CCT-beta | CCTB | MGC142074 | MGC142076 | PRO1633 | TCP-1-beta | chaperonin containing TCP1, subunit 2 (beta) | - | HPRD,BioGRID | 10748209|11580269 |
| CCT5 | CCT-epsilon | CCTE | KIAA0098 | TCP-1-epsilon | chaperonin containing TCP1, subunit 5 (epsilon) | - | HPRD,BioGRID | 11580269 |
| CCT8 | C21orf112 | Cctq | D21S246 | KIAA0002 | PRED71 | chaperonin containing TCP1, subunit 8 (theta) | - | HPRD | 10748209 |
| CFL1 | CFL | cofilin 1 (non-muscle) | Two-hybrid | BioGRID | 16189514 |
| CFL2 | NEM7 | cofilin 2 (muscle) | Affinity Capture-Western Two-hybrid | BioGRID | 16189514 |
| CIITA | C2TA | CIITAIV | MHC2TA | NLRA | class II, major histocompatibility complex, transactivator | Beta-actin interacts with p-MHC2TA. | BIND | 15502823 |
| CLIC4 | CLIC4L | DKFZp566G223 | FLJ38640 | H1 | MTCLIC | huH1 | p64H1 | chloride intracellular channel 4 | - | HPRD | 11563969 |
| CPNE1 | COPN1 | CPN1 | MGC1142 | copine I | - | HPRD | 12522145 |
| CPNE2 | COPN2 | CPN2 | DKFZp686E06199 | MGC16924 | copine II | - | HPRD | 12522145 |
| CPNE4 | COPN4 | CPN4 | MGC15604 | copine IV | - | HPRD | 12522145 |
| DNASE1 | DKFZp686H0155 | DNL1 | DRNI | FLJ38093 | FLJ44902 | deoxyribonuclease I | DNASE1 (DNase I) interacts with ACTB (beta-actin). | BIND | 15711563 |
| DSTN | ACTDP | ADF | bA462D18.2 | destrin (actin depolymerizing factor) | Two-hybrid | BioGRID | 16189514 |
| DYNLL1 | DLC1 | DLC8 | DNCL1 | DNCLC1 | LC8 | LC8a | MGC126137 | MGC126138 | PIN | hdlc1 | dynein, light chain, LC8-type 1 | - | HPRD | 14760703 |
| EMD | EDMD | LEMD5 | STA | emerin | - | HPRD | 12670476 |
| EZR | CVIL | CVL | DKFZp762H157 | FLJ26216 | MGC1584 | VIL2 | ezrin | - | HPRD | 7876308 |
| FLII | FLI | FLIL | Fli1 | MGC39265 | flightless I homolog (Drosophila) | - | HPRD | 9525888 |
| GSN | DKFZp313L0718 | gelsolin (amyloidosis, Finnish type) | - | HPRD | 10945978 |
| HNRNPU | HNRPU | SAF-A | U21.1 | heterogeneous nuclear ribonucleoprotein U (scaffold attachment factor A) | ACTB (beta-actin) interacts with HNRPU (hnRNP U). | BIND | 15711563 |
| IFI6 | 6-16 | FAM14C | G1P3 | IFI-6-16 | IFI616 | interferon, alpha-inducible protein 6 | Beta-actin interacts with p-G1P3. | BIND | 15502823 |
| LGALS13 | GAL13 | PLAC8 | PP13 | lectin, galactoside-binding, soluble, 13 | - | HPRD | 15009185 |
| MYC | bHLHe39 | c-Myc | v-myc myelocytomatosis viral oncogene homolog (avian) | Co-purification | BioGRID | 11839798 |
| NCALD | MGC33870 | MGC74858 | neurocalcin delta | Affinity Capture-Western | BioGRID | 16189514 |
| NCF1 | FLJ79451 | NCF1A | NOXO2 | SH3PXD1A | p47phox | neutrophil cytosolic factor 1 | Far Western | BioGRID | 11027608 |
| NCF1C | SH3PXD1C | neutrophil cytosolic factor 1C pseudogene | - | HPRD | 11027608 |
| NCF2 | FLJ93058 | NCF-2 | NOXA2 | P67-PHOX | P67PHOX | neutrophil cytosolic factor 2 | Far Western | BioGRID | 11027608 |
| NEBL | FLJ53769 | LNEBL | MGC119746 | MGC119747 | bA56H7.1 | nebulette | - | HPRD | 10470015 |
| NRAP | - | nebulin-related anchoring protein | - | HPRD | 9295142 |
| OSTF1 | FLJ20559 | OSF | SH3P2 | bA235O14.1 | osteoclast stimulating factor 1 | SH3P2 interacts with beta-actin. | BIND | 15135048 |
| P2RX7 | MGC20089 | P2X7 | purinergic receptor P2X, ligand-gated ion channel, 7 | - | HPRD | 11707406 |
| PCYT1B | CCT-beta | CTB | phosphate cytidylyltransferase 1, choline, beta | Co-crystal Structure | BioGRID | 11580269 |
| PFDN1 | PDF | PFD1 | prefoldin subunit 1 | - | HPRD | 11535601 |
| PFN1 | - | profilin 1 | - | HPRD,BioGRID | 10411937 |
| PFN2 | D3S1319E | PFL | profilin 2 | - | HPRD | 10600384 |
| PLD1 | - | phospholipase D1, phosphatidylcholine-specific | - | HPRD | 11373276 |
| PLD2 | - | phospholipase D2 | - | HPRD | 11373276 |
| POLR1B | FLJ10816 | FLJ21921 | MGC131780 | RPA135 | RPA2 | Rpo1-2 | polymerase (RNA) I polypeptide B, 128kDa | Pol I interacts with actin. | BIND | 15558034 |
| PTN | HARP | HBGF8 | HBNF | NEGF1 | pleiotrophin | - | HPRD | 9739080 |
| RAB8B | FLJ38125 | RAB8B, member RAS oncogene family | Affinity Capture-Western | BioGRID | 12639940 |
| RAC1 | MGC111543 | MIG5 | TC-25 | p21-Rac1 | ras-related C3 botulinum toxin substrate 1 (rho family, small GTP binding protein Rac1) | - | HPRD,BioGRID | 11027608 |
| RAC2 | EN-7 | Gx | HSPC022 | ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2) | - | HPRD,BioGRID | 11027608 |
| RIBIN | - | rRNA promoter binding protein | Actin interacts with rDNA. | BIND | 15558034 |
| RRN3 | DKFZp566E104 | MGC104238 | TIFIA | RRN3 RNA polymerase I transcription factor homolog (S. cerevisiae) | TIF-IA interacts with actin. | BIND | 15558034 |
| SMARCA4 | BAF190 | BRG1 | FLJ39786 | SNF2 | SNF2-BETA | SNF2L4 | SNF2LB | SWI2 | hSNF2b | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 | - | HPRD,BioGRID | 9845365 |
| SMARCC2 | BAF170 | CRACC2 | Rsc8 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 2 | Affinity Capture-Western | BioGRID | 9845365 |
| SMARCE1 | BAF57 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily e, member 1 | Affinity Capture-Western | BioGRID | 9845365 |
| SPTAN1 | (ALPHA)II-SPECTRIN | FLJ44613 | NEAS | spectrin, alpha, non-erythrocytic 1 (alpha-fodrin) | - | HPRD | 9209005 |
| SPTBN2 | GTRAP41 | SCA5 | spectrin, beta, non-erythrocytic 2 | Reconstituted Complex | BioGRID | 11357136 |
| SSH1 | FLJ38102 | KIAA1298 | SSH-1 | slingshot homolog 1 (Drosophila) | Reconstituted Complex | BioGRID | 11832213 |
| SSH2 | KIAA1725 | MGC78588 | SSH-2 | slingshot homolog 2 (Drosophila) | Reconstituted Complex | BioGRID | 11832213 |
| SYNJ2BP | ARIP2 | FLJ11271 | FLJ41973 | OMP25 | synaptojanin 2 binding protein | - | HPRD | 11673475 |11696420 |
| TANC1 | KIAA1728 | ROLSB | TANC | tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1 | - | HPRD | 15673434 |
| TPM2 | AMCD1 | DA1 | DA2B | TMSB | tropomyosin 2 (beta) | - | HPRD | 9108196 |
| TPM3 | FLJ41118 | MGC14582 | MGC3261 | MGC72094 | NEM1 | OK/SW-cl.5 | TM-5 | TM3 | TM30 | TM30nm | TPMsk3 | TRK | hscp30 | tropomyosin 3 | - | HPRD | 9108196 |
| XPO6 | EXP6 | FLJ22519 | KIAA0370 | RANBP20 | exportin 6 | - | HPRD | 14592989 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG FOCAL ADHESION | 201 | 138 | All SZGR 2.0 genes in this pathway |
| KEGG ADHERENS JUNCTION | 75 | 53 | All SZGR 2.0 genes in this pathway |
| KEGG TIGHT JUNCTION | 134 | 86 | All SZGR 2.0 genes in this pathway |
| KEGG LEUKOCYTE TRANSENDOTHELIAL MIGRATION | 118 | 78 | All SZGR 2.0 genes in this pathway |
| KEGG REGULATION OF ACTIN CYTOSKELETON | 216 | 144 | All SZGR 2.0 genes in this pathway |
| KEGG VIBRIO CHOLERAE INFECTION | 56 | 32 | All SZGR 2.0 genes in this pathway |
| KEGG PATHOGENIC ESCHERICHIA COLI INFECTION | 59 | 36 | All SZGR 2.0 genes in this pathway |
| KEGG HYPERTROPHIC CARDIOMYOPATHY HCM | 85 | 65 | All SZGR 2.0 genes in this pathway |
| KEGG ARRHYTHMOGENIC RIGHT VENTRICULAR CARDIOMYOPATHY ARVC | 76 | 59 | All SZGR 2.0 genes in this pathway |
| KEGG DILATED CARDIOMYOPATHY | 92 | 68 | All SZGR 2.0 genes in this pathway |
| KEGG VIRAL MYOCARDITIS | 73 | 58 | All SZGR 2.0 genes in this pathway |
| WNT SIGNALING | 89 | 71 | All SZGR 2.0 genes in this pathway |
| REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | 28 | 15 | All SZGR 2.0 genes in this pathway |
| REACTOME PROTEIN FOLDING | 53 | 30 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF PROTEINS | 518 | 242 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA UP | 783 | 507 | All SZGR 2.0 genes in this pathway |
| FOURNIER ACINAR DEVELOPMENT LATE DN | 21 | 13 | All SZGR 2.0 genes in this pathway |
| THUM SYSTOLIC HEART FAILURE UP | 423 | 283 | All SZGR 2.0 genes in this pathway |
| CASORELLI ACUTE PROMYELOCYTIC LEUKEMIA DN | 663 | 425 | All SZGR 2.0 genes in this pathway |
| TIEN INTESTINE PROBIOTICS 6HR UP | 55 | 34 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION UP | 1278 | 748 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS UP | 214 | 155 | All SZGR 2.0 genes in this pathway |
| WAMUNYOKOLI OVARIAN CANCER LMP UP | 265 | 158 | All SZGR 2.0 genes in this pathway |
| MAYBURD RESPONSE TO L663536 DN | 56 | 32 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY SERUM DEPRIVATION UP | 552 | 347 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND SERUM DEPRIVATION UP | 211 | 136 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN DN | 770 | 415 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA DN | 394 | 258 | All SZGR 2.0 genes in this pathway |
| CAVARD LIVER CANCER MALIGNANT VS BENIGN | 32 | 19 | All SZGR 2.0 genes in this pathway |
| RASHI RESPONSE TO IONIZING RADIATION 3 | 48 | 31 | All SZGR 2.0 genes in this pathway |
| DIRMEIER LMP1 RESPONSE EARLY | 66 | 48 | All SZGR 2.0 genes in this pathway |
| DACOSTA LOW DOSE UV RESPONSE VIA ERCC3 XPCS DN | 9 | 6 | All SZGR 2.0 genes in this pathway |
| JAZAG TGFB1 SIGNALING VIA SMAD4 UP | 108 | 66 | All SZGR 2.0 genes in this pathway |
| HATADA METHYLATED IN LUNG CANCER UP | 390 | 236 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS DN | 848 | 527 | All SZGR 2.0 genes in this pathway |
| RAHMAN TP53 TARGETS PHOSPHORYLATED | 21 | 18 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC TARGETS WITH EBOX | 230 | 156 | All SZGR 2.0 genes in this pathway |
| PETRETTO CARDIAC HYPERTROPHY | 34 | 26 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 7P22 AMPLICON | 38 | 27 | All SZGR 2.0 genes in this pathway |
| IIZUKA LIVER CANCER PROGRESSION G1 G2 DN | 25 | 18 | All SZGR 2.0 genes in this pathway |
| GNATENKO PLATELET SIGNATURE | 48 | 28 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS PLASMABLAST UP | 398 | 262 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT REJECTED VS OK UP | 87 | 58 | All SZGR 2.0 genes in this pathway |
| CHEN LUNG CANCER SURVIVAL | 28 | 22 | All SZGR 2.0 genes in this pathway |
| CHESLER BRAIN HIGHEST EXPRESSION | 40 | 29 | All SZGR 2.0 genes in this pathway |
| MAGRANGEAS MULTIPLE MYELOMA IGG VS IGA DN | 26 | 16 | All SZGR 2.0 genes in this pathway |
| FLECHNER PBL KIDNEY TRANSPLANT OK VS DONOR UP | 151 | 100 | All SZGR 2.0 genes in this pathway |
| YAO HOXA10 TARGETS VIA PROGESTERONE UP | 79 | 58 | All SZGR 2.0 genes in this pathway |
| MATSUDA NATURAL KILLER DIFFERENTIATION | 475 | 313 | All SZGR 2.0 genes in this pathway |
| GUO HEX TARGETS DN | 65 | 36 | All SZGR 2.0 genes in this pathway |
| MOREAUX MULTIPLE MYELOMA BY TACI UP | 412 | 249 | All SZGR 2.0 genes in this pathway |
| WEIGEL OXIDATIVE STRESS BY TBH AND H2O2 | 36 | 24 | All SZGR 2.0 genes in this pathway |
| KAAB FAILED HEART ATRIUM DN | 141 | 99 | All SZGR 2.0 genes in this pathway |
| JIANG AGING CEREBRAL CORTEX UP | 36 | 27 | All SZGR 2.0 genes in this pathway |
| HU GENOTOXIN ACTION DIRECT VS INDIRECT 4HR | 37 | 22 | All SZGR 2.0 genes in this pathway |
| MCCLUNG CREB1 TARGETS DN | 57 | 39 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 24HR DN | 148 | 102 | All SZGR 2.0 genes in this pathway |
| MODY HIPPOCAMPUS NEONATAL | 35 | 25 | All SZGR 2.0 genes in this pathway |
| KAYO AGING MUSCLE UP | 244 | 165 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 14HR DN | 298 | 200 | All SZGR 2.0 genes in this pathway |
| MACLACHLAN BRCA1 TARGETS DN | 16 | 12 | All SZGR 2.0 genes in this pathway |
| LEE CALORIE RESTRICTION MUSCLE UP | 43 | 33 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 UP | 428 | 266 | All SZGR 2.0 genes in this pathway |
| KEEN RESPONSE TO ROSIGLITAZONE DN | 106 | 68 | All SZGR 2.0 genes in this pathway |
| WHITESIDE CISPLATIN RESISTANCE DN | 13 | 9 | All SZGR 2.0 genes in this pathway |
| KAYO CALORIE RESTRICTION MUSCLE UP | 95 | 64 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 6HR UP | 71 | 48 | All SZGR 2.0 genes in this pathway |
| ZAMORA NOS2 TARGETS DN | 96 | 71 | All SZGR 2.0 genes in this pathway |
| JIANG AGING HYPOTHALAMUS UP | 47 | 31 | All SZGR 2.0 genes in this pathway |
| ZHONG SECRETOME OF LUNG CANCER AND MACROPHAGE | 77 | 50 | All SZGR 2.0 genes in this pathway |
| ZHONG SECRETOME OF LUNG CANCER AND ENDOTHELIUM | 66 | 47 | All SZGR 2.0 genes in this pathway |
| ZHONG SECRETOME OF LUNG CANCER AND FIBROBLAST | 132 | 93 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS UP | 673 | 430 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS UP | 602 | 364 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS UP | 601 | 369 | All SZGR 2.0 genes in this pathway |
| BASSO HAIRY CELL LEUKEMIA DN | 80 | 66 | All SZGR 2.0 genes in this pathway |
| LABBE WNT3A TARGETS UP | 112 | 71 | All SZGR 2.0 genes in this pathway |
| LABBE TGFB1 TARGETS UP | 102 | 64 | All SZGR 2.0 genes in this pathway |
| LABBE TARGETS OF TGFB1 AND WNT3A UP | 111 | 70 | All SZGR 2.0 genes in this pathway |
| HSIAO HOUSEKEEPING GENES | 389 | 245 | All SZGR 2.0 genes in this pathway |
| FOURNIER ACINAR DEVELOPMENT LATE 2 | 277 | 172 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 2 | 86 | 50 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 17 | 181 | 101 | All SZGR 2.0 genes in this pathway |
| SASSON RESPONSE TO GONADOTROPHINS UP | 91 | 59 | All SZGR 2.0 genes in this pathway |
| SASSON RESPONSE TO FORSKOLIN DN | 88 | 68 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS CONFLUENT | 567 | 365 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS UP | 504 | 321 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 2 UP | 140 | 94 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 UP | 430 | 288 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE LATE | 1137 | 655 | All SZGR 2.0 genes in this pathway |
| DORMOY ELAVL1 TARGETS | 17 | 10 | All SZGR 2.0 genes in this pathway |
| PURBEY TARGETS OF CTBP1 AND SATB1 UP | 87 | 52 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION DN | 1080 | 713 | All SZGR 2.0 genes in this pathway |
| GUO TARGETS OF IRS1 AND IRS2 | 98 | 67 | All SZGR 2.0 genes in this pathway |
| HUANG GATA2 TARGETS DN | 72 | 52 | All SZGR 2.0 genes in this pathway |
| STEINER ERYTHROCYTE MEMBRANE GENES | 15 | 10 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-124.1 | 412 | 418 | 1A | hsa-miR-124a | UUAAGGCACGCGGUGAAUGCCA |
| miR-145 | 145 | 152 | 1A,m8 | hsa-miR-145 | GUCCAGUUUUCCCAGGAAUCCCUU |
| miR-205 | 500 | 506 | m8 | hsa-miR-205 | UCCUUCAUUCCACCGGAGUCUG |
| miR-486 | 272 | 278 | 1A | hsa-miR-486 | UCCUGUACUGAGCUGCCCCGAG |
| miR-544 | 53 | 59 | 1A | hsa-miR-544 | AUUCUGCAUUUUUAGCAAGU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.