Gene Page: NOD2
Summary ?
| GeneID | 64127 |
| Symbol | NOD2 |
| Synonyms | ACUG|BLAU|CARD15|CD|CLR16.3|IBD1|NLRC2|NOD2B|PSORAS1 |
| Description | nucleotide binding oligomerization domain containing 2 |
| Reference | MIM:605956|HGNC:HGNC:5331|Ensembl:ENSG00000167207|HPRD:05810|Vega:OTTHUMG00000133171 |
| Gene type | protein-coding |
| Map location | 16q21 |
| Pascal p-value | 9.841E-4 |
| DMG | 1 (# studies) |
| eGene | Cerebellar Hemisphere Cerebellum Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Montano_2016 | Genome-wide DNA methylation analysis | This dataset includes 172 replicated associations between CpGs with schizophrenia. | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| GSMA_IIE | Genome scan meta-analysis (European-ancestry samples) | Psr: 0.01775 | |
| Expression | Meta-analysis of gene expression | P value: 1.735 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg04172533 | 16 | 50743027 | NOD2 | 9.43E-5 | -0.005 | 0.119 | DMG:Montano_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs1507047 | chr16 | 50932778 | NOD2 | 64127 | 0.01 | cis |
Section II. Transcriptome annotation
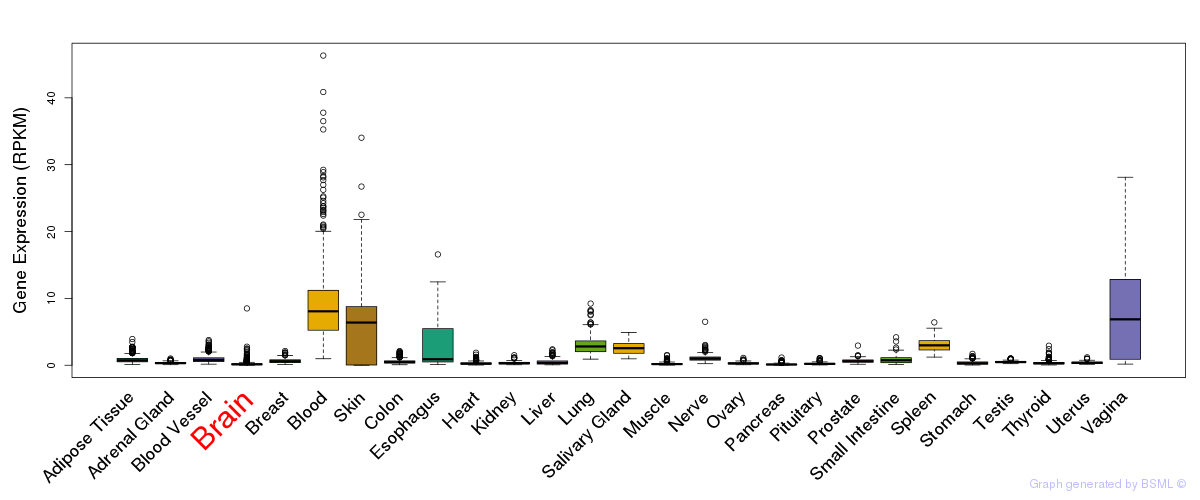
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0032500 | muramyl dipeptide binding | IDA | 15998797 | |
| GO:0019901 | protein kinase binding | IPI | 15075345 | |
| GO:0019899 | enzyme binding | IPI | 15753091 | |
| GO:0050700 | CARD domain binding | IPI | 11087742 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0002367 | cytokine production during immune response | IMP | 16260731 | |
| GO:0002224 | toll-like receptor signaling pathway | IEA | - | |
| GO:0016045 | detection of bacterium | IDA | 15653568 | |
| GO:0007242 | intracellular signaling cascade | IDA | 15653568 | |
| GO:0042981 | regulation of apoptosis | IEA | - | |
| GO:0051092 | positive regulation of NF-kappaB transcription factor activity | IDA | 11385577 | |
| GO:0042742 | defense response to bacterium | IDA | 15653568 | |
| GO:0032740 | positive regulation of interleukin-17 production | IMP | 17919942 | |
| GO:0051259 | protein oligomerization | TAS | 15967716 | |
| GO:0032498 | detection of muramyl dipeptide | IDA | 15998797 | |
| GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB cascade | IDA | 11087742 | |
| GO:0050718 | positive regulation of interleukin-1 beta secretion | IDA | 15107016 | |
| GO:0045087 | innate immune response | NAS | 17690884 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005829 | cytosol | IDA | 15998797 | |
| GO:0005622 | intracellular | IEA | - | |
| GO:0005737 | cytoplasm | TAS | 15967716 | |
| GO:0009986 | cell surface | IDA | 16714539 | |
| GO:0005886 | plasma membrane | IDA | 15998797 | |
| GO:0008180 | signalosome | IDA | 17337451 | |
| GO:0031982 | vesicle | IDA | 15998797 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG NOD LIKE RECEPTOR SIGNALING PATHWAY | 62 | 47 | All SZGR 2.0 genes in this pathway |
| PID NFKAPPAB CANONICAL PATHWAY | 23 | 20 | All SZGR 2.0 genes in this pathway |
| REACTOME TRIF MEDIATED TLR3 SIGNALING | 74 | 54 | All SZGR 2.0 genes in this pathway |
| REACTOME TAK1 ACTIVATES NFKB BY PHOSPHORYLATION AND ACTIVATION OF IKKS COMPLEX | 23 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME MAP KINASE ACTIVATION IN TLR CASCADE | 50 | 37 | All SZGR 2.0 genes in this pathway |
| REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | 16 | 9 | All SZGR 2.0 genes in this pathway |
| REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | 18 | 12 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY ILS | 107 | 86 | All SZGR 2.0 genes in this pathway |
| REACTOME IL1 SIGNALING | 39 | 30 | All SZGR 2.0 genes in this pathway |
| REACTOME TRAF6 MEDIATED INDUCTION OF NFKB AND MAP KINASES UPON TLR7 8 OR 9 ACTIVATION | 77 | 57 | All SZGR 2.0 genes in this pathway |
| REACTOME NFKB AND MAP KINASES ACTIVATION MEDIATED BY TLR4 SIGNALING REPERTOIRE | 72 | 53 | All SZGR 2.0 genes in this pathway |
| REACTOME MYD88 MAL CASCADE INITIATED ON PLASMA MEMBRANE | 83 | 63 | All SZGR 2.0 genes in this pathway |
| REACTOME INNATE IMMUNE SYSTEM | 279 | 178 | All SZGR 2.0 genes in this pathway |
| REACTOME ACTIVATED TLR4 SIGNALLING | 93 | 69 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME TOLL RECEPTOR CASCADES | 118 | 84 | All SZGR 2.0 genes in this pathway |
| REACTOME NOD1 2 SIGNALING PATHWAY | 30 | 22 | All SZGR 2.0 genes in this pathway |
| REACTOME CYTOKINE SIGNALING IN IMMUNE SYSTEM | 270 | 204 | All SZGR 2.0 genes in this pathway |
| REACTOME NUCLEOTIDE BINDING DOMAIN LEUCINE RICH REPEAT CONTAINING RECEPTOR NLR SIGNALING PATHWAYS | 46 | 33 | All SZGR 2.0 genes in this pathway |
| SMIRNOV CIRCULATING ENDOTHELIOCYTES IN CANCER UP | 158 | 103 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION HSC DN | 187 | 115 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE EPIDERMIS DN | 508 | 354 | All SZGR 2.0 genes in this pathway |
| GEORGES TARGETS OF MIR192 AND MIR215 | 893 | 528 | All SZGR 2.0 genes in this pathway |
| CERVERA SDHB TARGETS 1 UP | 118 | 66 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 2 | 473 | 224 | All SZGR 2.0 genes in this pathway |
| VALK AML CLUSTER 5 | 33 | 24 | All SZGR 2.0 genes in this pathway |
| VERHAAK GLIOBLASTOMA MESENCHYMAL | 216 | 130 | All SZGR 2.0 genes in this pathway |
| BHAT ESR1 TARGETS VIA AKT1 UP | 281 | 183 | All SZGR 2.0 genes in this pathway |
| PEDRIOLI MIR31 TARGETS DN | 418 | 245 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-378 | 437 | 443 | 1A | hsa-miR-378 | CUCCUGACUCCAGGUCCUGUGU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.