| KEGG ADIPOCYTOKINE SIGNALING PATHWAY
| 67 | 57 | All SZGR 2.0 genes in this pathway |
| KEGG PATHWAYS IN CANCER
| 328 | 259 | All SZGR 2.0 genes in this pathway |
| KEGG RENAL CELL CARCINOMA
| 70 | 60 | All SZGR 2.0 genes in this pathway |
| BIOCARTA VITCB PATHWAY
| 11 | 6 | All SZGR 2.0 genes in this pathway |
| PID HIF2PATHWAY
| 34 | 29 | All SZGR 2.0 genes in this pathway |
| PID MYC ACTIV PATHWAY
| 79 | 62 | All SZGR 2.0 genes in this pathway |
| PID HIF1 TFPATHWAY
| 66 | 52 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF VITAMINS AND COFACTORS
| 51 | 36 | All SZGR 2.0 genes in this pathway |
| REACTOME INTEGRATION OF ENERGY METABOLISM
| 120 | 84 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSMEMBRANE TRANSPORT OF SMALL MOLECULES
| 413 | 270 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF INSULIN SECRETION
| 93 | 65 | All SZGR 2.0 genes in this pathway |
| REACTOME SLC MEDIATED TRANSMEMBRANE TRANSPORT
| 241 | 157 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSPORT OF GLUCOSE AND OTHER SUGARS BILE SALTS AND ORGANIC ACIDS METAL IONS AND AMINE COMPOUNDS
| 89 | 54 | All SZGR 2.0 genes in this pathway |
| REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS
| 12 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME GLUCOSE TRANSPORT
| 38 | 29 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF CARBOHYDRATES
| 247 | 154 | All SZGR 2.0 genes in this pathway |
| WINTER HYPOXIA UP
| 92 | 57 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE LIVE DN
| 384 | 220 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP
| 1382 | 904 | All SZGR 2.0 genes in this pathway |
| WANG LMO4 TARGETS UP
| 372 | 227 | All SZGR 2.0 genes in this pathway |
| JAEGER METASTASIS DN
| 258 | 141 | All SZGR 2.0 genes in this pathway |
| ELVIDGE HYPOXIA UP
| 171 | 112 | All SZGR 2.0 genes in this pathway |
| ELVIDGE HYPOXIA BY DMOG UP
| 130 | 85 | All SZGR 2.0 genes in this pathway |
| ELVIDGE HIF1A TARGETS DN
| 91 | 58 | All SZGR 2.0 genes in this pathway |
| ELVIDGE HIF1A AND HIF2A TARGETS DN
| 104 | 72 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED UP
| 633 | 376 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC UP
| 722 | 443 | All SZGR 2.0 genes in this pathway |
| PROVENZANI METASTASIS DN
| 136 | 94 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE EPIDERMIS UP
| 293 | 179 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE DN
| 485 | 334 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY SERUM DEPRIVATION DN
| 234 | 147 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA UP
| 536 | 340 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 4 5WK UP
| 271 | 175 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL TGFB1 TARGETS UP
| 169 | 127 | All SZGR 2.0 genes in this pathway |
| RASHI RESPONSE TO IONIZING RADIATION 1
| 45 | 27 | All SZGR 2.0 genes in this pathway |
| MARTORIATI MDM4 TARGETS NEUROEPITHELIUM UP
| 176 | 111 | All SZGR 2.0 genes in this pathway |
| GRUETZMANN PANCREATIC CANCER UP
| 358 | 245 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK
| 1442 | 892 | All SZGR 2.0 genes in this pathway |
| GROSS HYPOXIA VIA ELK3 DN
| 156 | 106 | All SZGR 2.0 genes in this pathway |
| GROSS HYPOXIA VIA HIF1A DN
| 110 | 78 | All SZGR 2.0 genes in this pathway |
| GROSS HYPOXIA VIA ELK3 AND HIF1A UP
| 142 | 104 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS UP
| 1037 | 673 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS
| 957 | 597 | All SZGR 2.0 genes in this pathway |
| LIAO HAVE SOX4 BINDING SITES
| 40 | 26 | All SZGR 2.0 genes in this pathway |
| LIAO METASTASIS
| 539 | 324 | All SZGR 2.0 genes in this pathway |
| RICKMAN TUMOR DIFFERENTIATED WELL VS POORLY DN
| 382 | 224 | All SZGR 2.0 genes in this pathway |
| RICKMAN METASTASIS DN
| 261 | 155 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 12HR UP
| 116 | 79 | All SZGR 2.0 genes in this pathway |
| RICKMAN HEAD AND NECK CANCER C
| 113 | 47 | All SZGR 2.0 genes in this pathway |
| LE EGR2 TARGETS DN
| 108 | 84 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL AND BRAIN QTL TRANS
| 185 | 114 | All SZGR 2.0 genes in this pathway |
| WILLERT WNT SIGNALING
| 24 | 13 | All SZGR 2.0 genes in this pathway |
| GOLDRATH IMMUNE MEMORY
| 65 | 42 | All SZGR 2.0 genes in this pathway |
| NEMETH INFLAMMATORY RESPONSE LPS UP
| 88 | 64 | All SZGR 2.0 genes in this pathway |
| CHIARETTI ACUTE LYMPHOBLASTIC LEUKEMIA ZAP70
| 67 | 33 | All SZGR 2.0 genes in this pathway |
| RADMACHER AML PROGNOSIS
| 78 | 52 | All SZGR 2.0 genes in this pathway |
| SESTO RESPONSE TO UV C8
| 72 | 56 | All SZGR 2.0 genes in this pathway |
| KIM HYPOXIA
| 25 | 21 | All SZGR 2.0 genes in this pathway |
| HARRIS HYPOXIA
| 81 | 64 | All SZGR 2.0 genes in this pathway |
| KANG FLUOROURACIL RESISTANCE DN
| 16 | 10 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP
| 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| LEONARD HYPOXIA
| 47 | 35 | All SZGR 2.0 genes in this pathway |
| YAMAZAKI TCEB3 TARGETS DN
| 215 | 132 | All SZGR 2.0 genes in this pathway |
| SARTIPY NORMAL AT INSULIN RESISTANCE UP
| 34 | 27 | All SZGR 2.0 genes in this pathway |
| JIANG HYPOXIA NORMAL
| 311 | 205 | All SZGR 2.0 genes in this pathway |
| SEMENZA HIF1 TARGETS
| 36 | 32 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RESPONSE TO TRABECTEDIN DN
| 271 | 175 | All SZGR 2.0 genes in this pathway |
| DOUGLAS BMI1 TARGETS UP
| 566 | 371 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR DN
| 911 | 527 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR DN
| 1011 | 592 | All SZGR 2.0 genes in this pathway |
| GHO ATF5 TARGETS DN
| 16 | 10 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 STIMULATED
| 1022 | 619 | All SZGR 2.0 genes in this pathway |
| COATES MACROPHAGE M1 VS M2 DN
| 78 | 44 | All SZGR 2.0 genes in this pathway |
| ACEVEDO METHYLATED IN LIVER CANCER DN
| 940 | 425 | All SZGR 2.0 genes in this pathway |
| LABBE TARGETS OF TGFB1 AND WNT3A DN
| 108 | 68 | All SZGR 2.0 genes in this pathway |
| WINTER HYPOXIA METAGENE
| 242 | 168 | All SZGR 2.0 genes in this pathway |
| SHEDDEN LUNG CANCER POOR SURVIVAL A6
| 456 | 285 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA CHEMOTAXIS DN
| 457 | 302 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA HAPTOTAXIS DN
| 668 | 419 | All SZGR 2.0 genes in this pathway |
| VALK AML CLUSTER 7
| 28 | 16 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER SUBCLASS S1
| 237 | 159 | All SZGR 2.0 genes in this pathway |
| DANG REGULATED BY MYC UP
| 72 | 53 | All SZGR 2.0 genes in this pathway |
| WONG EMBRYONIC STEM CELL CORE
| 335 | 193 | All SZGR 2.0 genes in this pathway |
| WANG RESPONSE TO GSK3 INHIBITOR SB216763 DN
| 374 | 217 | All SZGR 2.0 genes in this pathway |
| QI HYPOXIA
| 140 | 96 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 24HR UP
| 324 | 193 | All SZGR 2.0 genes in this pathway |
| WANG ADIPOGENIC GENES REPRESSED BY SIRT1
| 28 | 21 | All SZGR 2.0 genes in this pathway |
| BHAT ESR1 TARGETS NOT VIA AKT1 UP
| 211 | 131 | All SZGR 2.0 genes in this pathway |
| BHAT ESR1 TARGETS VIA AKT1 UP
| 281 | 183 | All SZGR 2.0 genes in this pathway |
| VANDESLUIS COMMD1 TARGETS GROUP 4 UP
| 19 | 12 | All SZGR 2.0 genes in this pathway |
| KASLER HDAC7 TARGETS 1 DN
| 17 | 13 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS UP
| 682 | 433 | All SZGR 2.0 genes in this pathway |
| HUANG GATA2 TARGETS DN
| 72 | 52 | All SZGR 2.0 genes in this pathway |
| PLASARI TGFB1 TARGETS 10HR UP
| 199 | 143 | All SZGR 2.0 genes in this pathway |
| KRIEG HYPOXIA NOT VIA KDM3A
| 770 | 480 | All SZGR 2.0 genes in this pathway |
| ZWANG CLASS 3 TRANSIENTLY INDUCED BY EGF
| 222 | 159 | All SZGR 2.0 genes in this pathway |
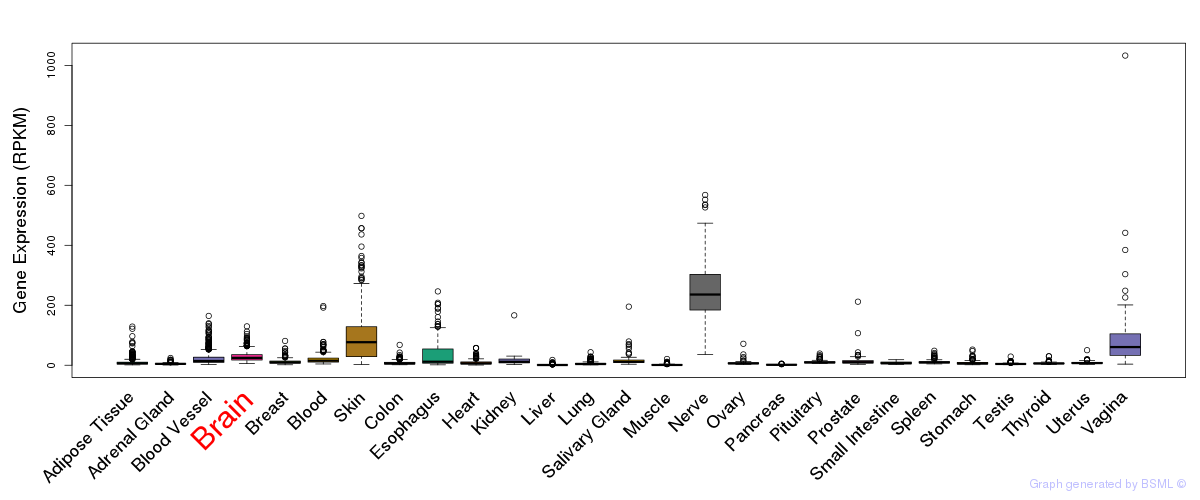
 Differentially methylated gene
Differentially methylated gene eQTL annotation
eQTL annotation