Gene Page: SOD1
Summary ?
| GeneID | 6647 |
| Symbol | SOD1 |
| Synonyms | ALS|ALS1|HEL-S-44|IPOA|SOD|hSod1|homodimer |
| Description | superoxide dismutase 1, soluble |
| Reference | MIM:147450|HGNC:HGNC:11179|Ensembl:ENSG00000142168|HPRD:00937|Vega:OTTHUMG00000084878 |
| Gene type | protein-coding |
| Map location | 21q22.11 |
| Pascal p-value | 0.31 |
| Sherlock p-value | 0.043 |
| Fetal beta | -0.452 |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 5 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs7132043 | chr12 | 80968399 | SOD1 | 6647 | 0.2 | trans |
Section II. Transcriptome annotation
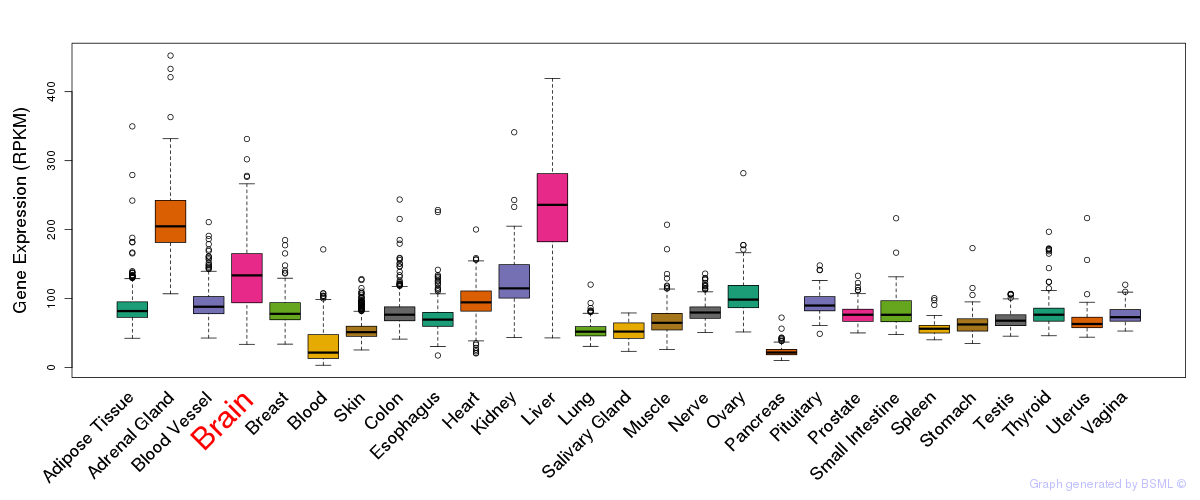
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| GAK | 0.89 | 0.88 |
| ARFGAP1 | 0.88 | 0.86 |
| DNAJC11 | 0.86 | 0.83 |
| CUL9 | 0.86 | 0.85 |
| PLA2G6 | 0.86 | 0.83 |
| ATG2A | 0.86 | 0.85 |
| ZNF839 | 0.86 | 0.82 |
| C16orf58 | 0.85 | 0.81 |
| DDX51 | 0.85 | 0.85 |
| RAB11FIP3 | 0.85 | 0.85 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.21 | -0.78 | -0.67 |
| MT-CO2 | -0.70 | -0.60 |
| AF347015.31 | -0.70 | -0.61 |
| AF347015.8 | -0.67 | -0.58 |
| C1orf54 | -0.67 | -0.62 |
| MT-CYB | -0.65 | -0.55 |
| CLEC2B | -0.65 | -0.60 |
| AF347015.27 | -0.64 | -0.57 |
| AF347015.33 | -0.64 | -0.53 |
| NOSTRIN | -0.63 | -0.50 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005507 | copper ion binding | IDA | 17008312 | |
| GO:0004784 | superoxide dismutase activity | IDA | 15544046 |17324120 | |
| GO:0008270 | zinc ion binding | IDA | 17381088 | |
| GO:0016209 | antioxidant activity | IEA | - | |
| GO:0016491 | oxidoreductase activity | IEA | - | |
| GO:0051087 | chaperone binding | IPI | 9726962 | |
| GO:0042803 | protein homodimerization activity | NAS | 9726962 |10837872 | |
| GO:0030346 | protein phosphatase 2B binding | IDA | 17324120 | |
| GO:0046872 | metal ion binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0048678 | response to axon injury | ISS | axon (GO term level: 5) | - |
| GO:0019226 | transmission of nerve impulse | ISS | neuron (GO term level: 5) | - |
| GO:0043524 | negative regulation of neuron apoptosis | ISS | neuron (GO term level: 9) | - |
| GO:0000187 | activation of MAPK activity | ISS | - | |
| GO:0000303 | response to superoxide | IDA | 16790527 | |
| GO:0001541 | ovarian follicle development | ISS | - | |
| GO:0002262 | myeloid cell homeostasis | ISS | - | |
| GO:0001895 | retina homeostasis | ISS | - | |
| GO:0001890 | placenta development | NAS | 12485882 | |
| GO:0001819 | positive regulation of cytokine production | IDA | 15544046 | |
| GO:0006302 | double-strand break repair | ISS | - | |
| GO:0006309 | DNA fragmentation during apoptosis | ISS | - | |
| GO:0008217 | regulation of blood pressure | ISS | - | |
| GO:0007283 | spermatogenesis | ISS | - | |
| GO:0009408 | response to heat | ISS | - | |
| GO:0007626 | locomotory behavior | ISS | - | |
| GO:0007605 | sensory perception of sound | ISS | - | |
| GO:0007569 | cell aging | IEP | 15377661 | |
| GO:0007569 | cell aging | IMP | 12871978 | |
| GO:0007566 | embryo implantation | ISS | - | |
| GO:0007566 | embryo implantation | NAS | 10920331 | |
| GO:0006749 | glutathione metabolic process | ISS | - | |
| GO:0006879 | cellular iron ion homeostasis | ISS | - | |
| GO:0043065 | positive regulation of apoptosis | IC | 16790527 | |
| GO:0033081 | regulation of T cell differentiation in the thymus | NAS | 16716898 | |
| GO:0042493 | response to drug | ISS | - | |
| GO:0042554 | superoxide release | IEA | - | |
| GO:0032287 | myelin maintenance in the peripheral nervous system | ISS | - | |
| GO:0043085 | positive regulation of catalytic activity | IDA | 17324120 | |
| GO:0019430 | removal of superoxide radicals | ISS | - | |
| GO:0042542 | response to hydrogen peroxide | ISS | - | |
| GO:0040014 | regulation of multicellular organism growth | ISS | - | |
| GO:0045541 | negative regulation of cholesterol biosynthetic process | IDA | 15473258 | |
| GO:0045859 | regulation of protein kinase activity | IDA | 16254550 | |
| GO:0050665 | hydrogen peroxide biosynthetic process | IDA | 15544046 | |
| GO:0050665 | hydrogen peroxide biosynthetic process | ISS | - | |
| GO:0046620 | regulation of organ growth | NAS | 16716898 | |
| GO:0046716 | muscle maintenance | ISS | - | |
| GO:0048538 | thymus development | NAS | 16716898 | |
| GO:0060088 | auditory receptor cell stereocilium organization | ISS | - | |
| GO:0060087 | relaxation of vascular smooth muscle | ISS | - | |
| GO:0045471 | response to ethanol | ISS | - | |
| GO:0055114 | oxidation reduction | IEA | - | |
| GO:0060047 | heart contraction | IDA | 9539776 | |
| GO:0060052 | neurofilament cytoskeleton organization | ISS | - | |
| GO:0051881 | regulation of mitochondrial membrane potential | IMP | 16790527 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0043025 | cell soma | IDA | axon, dendrite (GO term level: 4) | 17324120 |
| GO:0032839 | dendrite cytoplasm | IDA | dendrite (GO term level: 9) | 17324120 |
| GO:0005829 | cytosol | IDA | 16790527 | |
| GO:0005615 | extracellular space | IDA | 7172448 |9453566 | |
| GO:0005634 | nucleus | IDA | 9726962 |17504823 | |
| GO:0005737 | cytoplasm | IDA | 9726962 |11527942 |17077646 |17504823 | |
| GO:0005739 | mitochondrion | IDA | 16790527 | |
| GO:0005759 | mitochondrial matrix | NAS | 17008312 | |
| GO:0005777 | peroxisome | NAS | 15766328 | |
| GO:0005886 | plasma membrane | IDA | 17077646 |17324120 | |
| GO:0031012 | extracellular matrix | IDA | 9699963 | |
| GO:0031410 | cytoplasmic vesicle | IDA | 17077646 | |
| GO:0043234 | protein complex | IDA | 17324120 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG PEROXISOME | 78 | 47 | All SZGR 2.0 genes in this pathway |
| KEGG AMYOTROPHIC LATERAL SCLEROSIS ALS | 53 | 43 | All SZGR 2.0 genes in this pathway |
| KEGG HUNTINGTONS DISEASE | 185 | 109 | All SZGR 2.0 genes in this pathway |
| KEGG PRION DISEASES | 35 | 28 | All SZGR 2.0 genes in this pathway |
| BIOCARTA FREE PATHWAY | 10 | 7 | All SZGR 2.0 genes in this pathway |
| BIOCARTA LONGEVITY PATHWAY | 15 | 13 | All SZGR 2.0 genes in this pathway |
| PID ERA GENOMIC PATHWAY | 65 | 37 | All SZGR 2.0 genes in this pathway |
| PID HNF3A PATHWAY | 44 | 29 | All SZGR 2.0 genes in this pathway |
| REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | 89 | 56 | All SZGR 2.0 genes in this pathway |
| REACTOME HEMOSTASIS | 466 | 331 | All SZGR 2.0 genes in this pathway |
| REACTOME PLATELET ACTIVATION SIGNALING AND AGGREGATION | 208 | 138 | All SZGR 2.0 genes in this pathway |
| CHANDRAN METASTASIS TOP50 UP | 38 | 27 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| PROVENZANI METASTASIS DN | 136 | 94 | All SZGR 2.0 genes in this pathway |
| DARWICHE SKIN TUMOR PROMOTER UP | 142 | 96 | All SZGR 2.0 genes in this pathway |
| DARWICHE PAPILLOMA RISK LOW DN | 165 | 107 | All SZGR 2.0 genes in this pathway |
| DARWICHE PAPILLOMA RISK HIGH DN | 180 | 110 | All SZGR 2.0 genes in this pathway |
| DARWICHE SQUAMOUS CELL CARCINOMA UP | 146 | 104 | All SZGR 2.0 genes in this pathway |
| HU ANGIOGENESIS DN | 37 | 25 | All SZGR 2.0 genes in this pathway |
| AMUNDSON RESPONSE TO ARSENITE | 217 | 143 | All SZGR 2.0 genes in this pathway |
| GRUETZMANN PANCREATIC CANCER UP | 358 | 245 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| FRIDMAN SENESCENCE UP | 77 | 60 | All SZGR 2.0 genes in this pathway |
| GOTZMANN EPITHELIAL TO MESENCHYMAL TRANSITION DN | 206 | 136 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS | 775 | 494 | All SZGR 2.0 genes in this pathway |
| HOUSTIS ROS | 36 | 29 | All SZGR 2.0 genes in this pathway |
| ROSS AML OF FAB M7 TYPE | 68 | 44 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT REJECTED VS OK DN | 546 | 351 | All SZGR 2.0 genes in this pathway |
| HSIAO LIVER SPECIFIC GENES | 244 | 154 | All SZGR 2.0 genes in this pathway |
| BROWN MYELOID CELL DEVELOPMENT DN | 129 | 86 | All SZGR 2.0 genes in this pathway |
| NADLER OBESITY DN | 48 | 34 | All SZGR 2.0 genes in this pathway |
| FAELT B CLL WITH VH REARRANGEMENTS DN | 48 | 26 | All SZGR 2.0 genes in this pathway |
| TAKAO RESPONSE TO UVB RADIATION UP | 86 | 55 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RESPONSE TO TRABECTEDIN UP | 71 | 49 | All SZGR 2.0 genes in this pathway |
| NATSUME RESPONSE TO INTERFERON BETA DN | 52 | 33 | All SZGR 2.0 genes in this pathway |
| ZHANG PROLIFERATING VS QUIESCENT | 51 | 41 | All SZGR 2.0 genes in this pathway |
| HEDENFALK BREAST CANCER BRCA1 VS BRCA2 | 163 | 113 | All SZGR 2.0 genes in this pathway |
| WHITESIDE CISPLATIN RESISTANCE DN | 13 | 9 | All SZGR 2.0 genes in this pathway |
| XU GH1 AUTOCRINE TARGETS UP | 268 | 157 | All SZGR 2.0 genes in this pathway |
| VISALA RESPONSE TO HEAT SHOCK AND AGING DN | 14 | 12 | All SZGR 2.0 genes in this pathway |
| JIANG AGING HYPOTHALAMUS UP | 47 | 31 | All SZGR 2.0 genes in this pathway |
| HILLION HMGA1 TARGETS | 90 | 71 | All SZGR 2.0 genes in this pathway |
| HILLION HMGA1B TARGETS | 92 | 68 | All SZGR 2.0 genes in this pathway |
| KYNG ENVIRONMENTAL STRESS RESPONSE NOT BY GAMMA IN WS | 33 | 22 | All SZGR 2.0 genes in this pathway |
| KYNG ENVIRONMENTAL STRESS RESPONSE NOT BY UV IN WS | 12 | 9 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE DN | 195 | 135 | All SZGR 2.0 genes in this pathway |
| KYNG ENVIRONMENTAL STRESS RESPONSE UP | 56 | 41 | All SZGR 2.0 genes in this pathway |
| LEE METASTASIS AND ALTERNATIVE SPLICING DN | 45 | 31 | All SZGR 2.0 genes in this pathway |
| IWANAGA CARCINOGENESIS BY KRAS PTEN DN | 353 | 226 | All SZGR 2.0 genes in this pathway |
| ACEVEDO NORMAL TISSUE ADJACENT TO LIVER TUMOR DN | 354 | 216 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER DN | 540 | 340 | All SZGR 2.0 genes in this pathway |
| MATZUK PREOVULATORY FOLLICLE | 10 | 8 | All SZGR 2.0 genes in this pathway |
| CHUNG BLISTER CYTOTOXICITY UP | 134 | 84 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| HINATA NFKB TARGETS KERATINOCYTE UP | 91 | 63 | All SZGR 2.0 genes in this pathway |
| HINATA NFKB TARGETS FIBROBLAST UP | 84 | 60 | All SZGR 2.0 genes in this pathway |
| HSIAO HOUSEKEEPING GENES | 389 | 245 | All SZGR 2.0 genes in this pathway |
| YAMASHITA LIVER CANCER STEM CELL DN | 76 | 51 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER SUBCLASS S3 | 266 | 180 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS OLIGODENDROCYTE NUMBER CORR UP | 756 | 494 | All SZGR 2.0 genes in this pathway |
| KIM BIPOLAR DISORDER OLIGODENDROCYTE DENSITY CORR UP | 682 | 440 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG BOUND 8D | 658 | 397 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-377 | 107 | 113 | m8 | hsa-miR-377 | AUCACACAAAGGCAACUUUUGU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.