Gene Page: SPTAN1
Summary ?
| GeneID | 6709 |
| Symbol | SPTAN1 |
| Synonyms | EIEE5|NEAS|SPTA2 |
| Description | spectrin alpha, non-erythrocytic 1 |
| Reference | MIM:182810|HGNC:HGNC:11273|Ensembl:ENSG00000197694|HPRD:01684|Vega:OTTHUMG00000020754 |
| Gene type | protein-coding |
| Map location | 9q34.11 |
| Pascal p-value | 0.129 |
| Sherlock p-value | 0.868 |
| Fetal beta | 0.358 |
| eGene | Meta |
| Support | STRUCTURAL PLASTICITY G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS CompositeSet Darnell FMRP targets Ascano FMRP targets Potential synaptic genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.7272 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
General gene expression (GTEx)
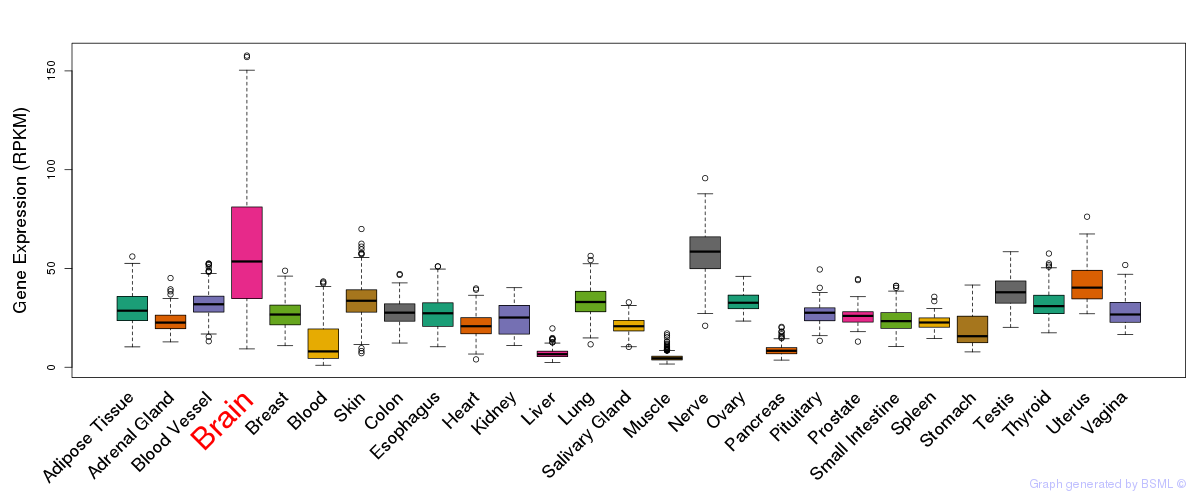
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| HNRNPUL1 | 0.97 | 0.94 |
| RAF1 | 0.97 | 0.96 |
| MTA2 | 0.97 | 0.94 |
| DDX23 | 0.97 | 0.97 |
| UBE2I | 0.97 | 0.95 |
| SMARCB1 | 0.97 | 0.94 |
| SF3B3 | 0.96 | 0.96 |
| TRA2B | 0.96 | 0.95 |
| WDR33 | 0.96 | 0.96 |
| FUBP1 | 0.96 | 0.96 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.31 | -0.78 | -0.92 |
| AF347015.27 | -0.78 | -0.92 |
| C5orf53 | -0.77 | -0.78 |
| MT-CO2 | -0.77 | -0.92 |
| HLA-F | -0.77 | -0.79 |
| AIFM3 | -0.76 | -0.80 |
| AF347015.33 | -0.76 | -0.90 |
| S100B | -0.75 | -0.84 |
| FXYD1 | -0.75 | -0.89 |
| MT-CYB | -0.75 | -0.90 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003779 | actin binding | TAS | 2307671 | |
| GO:0005509 | calcium ion binding | IEA | - | |
| GO:0005516 | calmodulin binding | IEA | - | |
| GO:0005200 | structural constituent of cytoskeleton | TAS | 2307671 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0051016 | barbed-end actin filament capping | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005829 | cytosol | EXP | 9624143 | |
| GO:0005856 | cytoskeleton | IEA | - | |
| GO:0005624 | membrane fraction | TAS | 2307671 | |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0008091 | spectrin | TAS | 2307671 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ABL1 | ABL | JTK7 | bcr/abl | c-ABL | p150 | v-abl | c-abl oncogene 1, receptor tyrosine kinase | Reconstituted Complex Two-hybrid | BioGRID | 9593709 |
| ACP1 | HAAP | MGC111030 | MGC3499 | acid phosphatase 1, soluble | - | HPRD | 11971983 |
| ACTB | PS1TP5BP1 | actin, beta | - | HPRD | 9209005 |
| ADD1 | ADDA | MGC3339 | MGC44427 | adducin 1 (alpha) | - | HPRD,BioGRID | 8663089 |
| ADD2 | ADDB | adducin 2 (beta) | - | HPRD | 7642559 |
| ANK1 | ANK | SPH1 | SPH2 | ankyrin 1, erythrocytic | - | HPRD | 492324 |2141335 |2970468 |2971657 |
| CAPN3 | CANP3 | CANPL3 | LGMD2 | LGMD2A | MGC10767 | MGC11121 | MGC14344 | MGC4403 | nCL-1 | p94 | calpain 3, (p94) | - | HPRD | 9642272 |
| CTNNA1 | CAP102 | FLJ36832 | catenin (cadherin-associated protein), alpha 1, 102kDa | - | HPRD | 11069925 |
| DDX24 | - | DEAD (Asp-Glu-Ala-Asp) box polypeptide 24 | Two-hybrid | BioGRID | 16169070 |
| DES | CMD1I | CSM1 | CSM2 | FLJ12025 | FLJ39719 | FLJ41013 | FLJ41793 | desmin | - | HPRD,BioGRID | 2939097 |
| DISC1 | C1orf136 | FLJ13381 | FLJ21640 | FLJ25311 | FLJ41105 | KIAA0457 | SCZD9 | disrupted in schizophrenia 1 | An unspecified isoform of DISC1 interacts with SPTAN1. | BIND | 14623284 |
| EPB41 | 4.1R | EL1 | HE | erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked) | Reconstituted Complex | BioGRID | 12044158 |
| EPB41 | 4.1R | EL1 | HE | erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked) | - | HPRD | 12049649 |
| EPB42 | MGC116735 | MGC116737 | PA | erythrocyte membrane protein band 4.2 | - | HPRD,BioGRID | 12049649 |
| ERCC4 | RAD1 | XPF | excision repair cross-complementing rodent repair deficiency, complementation group 4 | Affinity Capture-Western | BioGRID | 12571280 |
| EVL | RNB6 | Enah/Vasp-like | AlphaII-spectrin interacts with EVL. | BIND | 15656790 |
| FANCA | FA | FA-H | FA1 | FAA | FACA | FAH | FANCH | MGC75158 | Fanconi anemia, complementation group A | Affinity Capture-Western Co-purification | BioGRID | 10551855 |11401546 |12571280 |
| FANCC | FA3 | FAC | FACC | FLJ14675 | Fanconi anemia, complementation group C | Affinity Capture-Western Co-purification | BioGRID | 10551855 |11401546 |
| FANCG | FAG | XRCC9 | Fanconi anemia, complementation group G | Co-purification | BioGRID | 11401546 |
| GAP43 | B-50 | PP46 | growth associated protein 43 | - | HPRD | 10521589 |
| GRIA2 | GLUR2 | GLURB | GluR-K2 | HBGR2 | glutamate receptor, ionotropic, AMPA 2 | Affinity Capture-Western Far Western | BioGRID | 10576550 |
| GRIN1 | NMDA1 | NMDAR1 | NR1 | glutamate receptor, ionotropic, N-methyl D-aspartate 1 | - | HPRD | 9670010 |10862698 |
| MAP3K7IP2 | FLJ21885 | KIAA0733 | TAB2 | mitogen-activated protein kinase kinase kinase 7 interacting protein 2 | - | HPRD | 14743216 |
| NEFL | CMT1F | CMT2E | FLJ53642 | NF-L | NF68 | NFL | neurofilament, light polypeptide | - | HPRD,BioGRID | 1902666 |3121319 |
| PDE4D | DPDE3 | HSPDE4D | PDE4DN2 | STRK1 | phosphodiesterase 4D, cAMP-specific (phosphodiesterase E3 dunce homolog, Drosophila) | PDE4D4 interacts with Fodrin. This interaction was modelled on a demonstrated interaction between human PDE4D4 and Fodrin from an unspecified species. | BIND | 10571082 |
| PIN4 | EPVH | MGC138486 | PAR14 | PAR17 | protein (peptidylprolyl cis/trans isomerase) NIMA-interacting, 4 (parvulin) | Two-hybrid | BioGRID | 16169070 |
| PLEC1 | EBS1 | EBSO | HD1 | PCN | PLEC1b | PLTN | plectin 1, intermediate filament binding protein 500kDa | - | HPRD,BioGRID | 3027087 |
| PRKCB | MGC41878 | PKC-beta | PKCB | PRKCB1 | PRKCB2 | protein kinase C, beta | - | HPRD | 8163551 |
| PTOV1 | ACID2 | DKFZp586I111 | MGC71475 | prostate tumor overexpressed 1 | Two-hybrid | BioGRID | 16169070 |
| PTP4A1 | DKFZp779M0721 | HH72 | PRL-1 | PRL1 | PTP(CAAX1) | PTPCAAX1 | protein tyrosine phosphatase type IVA, member 1 | - | HPRD | 11971983 |
| SHANK1 | SPANK-1 | SSTRIP | synamon | SH3 and multiple ankyrin repeat domains 1 | - | HPRD,BioGRID | 11509555 |
| SHANK3 | KIAA1650 | PROSAP2 | PSAP2 | SPANK-2 | SH3 and multiple ankyrin repeat domains 3 | Two-hybrid | BioGRID | 11509555 |
| SLC9A2 | NHE2 | solute carrier family 9 (sodium/hydrogen exchanger), member 2 | - | HPRD | 10579058 |
| SPTB | HSpTB1 | spectrin, beta, erythrocytic | - | HPRD,BioGRID | 9890967 |
| SYN1 | SYN1a | SYN1b | SYNI | synapsin I | - | HPRD | 1905928 |
| TANC1 | KIAA1728 | ROLSB | TANC | tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1 | - | HPRD | 15673434 |
| TES | DKFZp586B2022 | MGC1146 | TESS | TESS-2 | testis derived transcript (3 LIM domains) | AlphaII-spectrin interacts with Tes. | BIND | 15656790 |
| TSSC4 | - | tumor suppressing subtransferable candidate 4 | Two-hybrid | BioGRID | 16169070 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG TIGHT JUNCTION | 134 | 86 | All SZGR 2.0 genes in this pathway |
| BIOCARTA FAS PATHWAY | 30 | 22 | All SZGR 2.0 genes in this pathway |
| BIOCARTA HIVNEF PATHWAY | 58 | 43 | All SZGR 2.0 genes in this pathway |
| BIOCARTA DEATH PATHWAY | 33 | 24 | All SZGR 2.0 genes in this pathway |
| BIOCARTA TNFR1 PATHWAY | 29 | 21 | All SZGR 2.0 genes in this pathway |
| BIOCARTA UCALPAIN PATHWAY | 18 | 11 | All SZGR 2.0 genes in this pathway |
| PID CASPASE PATHWAY | 52 | 39 | All SZGR 2.0 genes in this pathway |
| REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | 40 | 26 | All SZGR 2.0 genes in this pathway |
| REACTOME DEVELOPMENTAL BIOLOGY | 396 | 292 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CELL COMMUNICATION | 120 | 77 | All SZGR 2.0 genes in this pathway |
| REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | 13 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME AXON GUIDANCE | 251 | 188 | All SZGR 2.0 genes in this pathway |
| REACTOME NCAM SIGNALING FOR NEURITE OUT GROWTH | 64 | 49 | All SZGR 2.0 genes in this pathway |
| REACTOME L1CAM INTERACTIONS | 86 | 62 | All SZGR 2.0 genes in this pathway |
| REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | 23 | 19 | All SZGR 2.0 genes in this pathway |
| REACTOME NEPHRIN INTERACTIONS | 20 | 15 | All SZGR 2.0 genes in this pathway |
| REACTOME APOPTOSIS | 148 | 94 | All SZGR 2.0 genes in this pathway |
| REACTOME APOPTOTIC EXECUTION PHASE | 54 | 37 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA UP | 783 | 507 | All SZGR 2.0 genes in this pathway |
| VECCHI GASTRIC CANCER EARLY UP | 430 | 232 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 6HR DN | 41 | 29 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN NPM1 MUTATED SIGNATURE 1 DN | 126 | 86 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN NPM1 SIGNATURE 3 DN | 162 | 116 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION MONOCYTE UP | 204 | 140 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION ERYTHROCYTE UP | 157 | 104 | All SZGR 2.0 genes in this pathway |
| UDAYAKUMAR MED1 TARGETS DN | 240 | 171 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION DN | 329 | 219 | All SZGR 2.0 genes in this pathway |
| HAHTOLA MYCOSIS FUNGOIDES SKIN UP | 177 | 113 | All SZGR 2.0 genes in this pathway |
| KOKKINAKIS METHIONINE DEPRIVATION 48HR UP | 128 | 95 | All SZGR 2.0 genes in this pathway |
| KOKKINAKIS METHIONINE DEPRIVATION 96HR UP | 117 | 84 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE EPIDERMIS DN | 508 | 354 | All SZGR 2.0 genes in this pathway |
| CHIARADONNA NEOPLASTIC TRANSFORMATION KRAS DN | 142 | 95 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN DN | 770 | 415 | All SZGR 2.0 genes in this pathway |
| LANDIS ERBB2 BREAST TUMORS 324 UP | 150 | 93 | All SZGR 2.0 genes in this pathway |
| BARRIER CANCER RELAPSE NORMAL SAMPLE DN | 31 | 19 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA DN | 394 | 258 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE UP | 134 | 93 | All SZGR 2.0 genes in this pathway |
| WANG HCP PROSTATE CANCER | 111 | 69 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER MYC E2F1 UP | 56 | 34 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER E2F1 UP | 62 | 35 | All SZGR 2.0 genes in this pathway |
| LENAOUR DENDRITIC CELL MATURATION UP | 114 | 84 | All SZGR 2.0 genes in this pathway |
| ULE SPLICING VIA NOVA2 | 43 | 38 | All SZGR 2.0 genes in this pathway |
| APRELIKOVA BRCA1 TARGETS | 49 | 33 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS 8 | 86 | 57 | All SZGR 2.0 genes in this pathway |
| KAYO CALORIE RESTRICTION MUSCLE UP | 95 | 64 | All SZGR 2.0 genes in this pathway |
| BILD MYC ONCOGENIC SIGNATURE | 206 | 117 | All SZGR 2.0 genes in this pathway |
| RIZKI TUMOR INVASIVENESS 2D UP | 69 | 46 | All SZGR 2.0 genes in this pathway |
| ACEVEDO NORMAL TISSUE ADJACENT TO LIVER TUMOR DN | 354 | 216 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER TUMOR VS NORMAL ADJACENT TISSUE UP | 863 | 514 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER LUMINAL A UP | 84 | 52 | All SZGR 2.0 genes in this pathway |
| MALONEY RESPONSE TO 17AAG UP | 41 | 26 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS DN | 435 | 289 | All SZGR 2.0 genes in this pathway |
| CHEN METABOLIC SYNDROM NETWORK | 1210 | 725 | All SZGR 2.0 genes in this pathway |
| DANG REGULATED BY MYC DN | 253 | 192 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS OLIGODENDROCYTE NUMBER CORR UP | 756 | 494 | All SZGR 2.0 genes in this pathway |
| WANG RESPONSE TO GSK3 INHIBITOR SB216763 UP | 397 | 206 | All SZGR 2.0 genes in this pathway |
| CHYLA CBFA2T3 TARGETS UP | 387 | 225 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 2 UP | 140 | 94 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-103/107 | 48 | 54 | 1A | hsa-miR-103brain | AGCAGCAUUGUACAGGGCUAUGA |
| hsa-miR-107brain | AGCAGCAUUGUACAGGGCUAUCA | ||||
| miR-29 | 132 | 138 | m8 | hsa-miR-29aSZ | UAGCACCAUCUGAAAUCGGUU |
| hsa-miR-29bSZ | UAGCACCAUUUGAAAUCAGUGUU | ||||
| hsa-miR-29cSZ | UAGCACCAUUUGAAAUCGGU | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.