Gene Page: SRC
Summary ?
| GeneID | 6714 |
| Symbol | SRC |
| Synonyms | ASV|SRC1|c-SRC|p60-Src |
| Description | SRC proto-oncogene, non-receptor tyrosine kinase |
| Reference | MIM:190090|HGNC:HGNC:11283|Ensembl:ENSG00000197122|HPRD:01819|Vega:OTTHUMG00000032417 |
| Gene type | protein-coding |
| Map location | 20q12-q13 |
| Pascal p-value | 0.228 |
| Sherlock p-value | 0.968 |
| Fetal beta | 0.082 |
| Support | METABOTROPIC GLUTAMATE RECEPTOR G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS G2Cdb.humanNRC CompositeSet Potential synaptic genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 14.4947 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
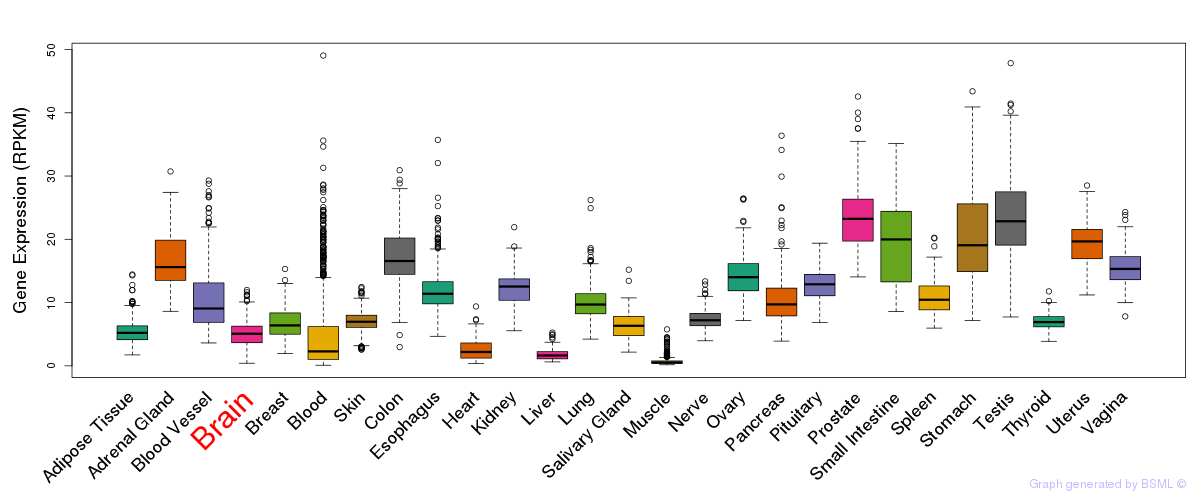
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| RASAL1 | 0.79 | 0.78 |
| RASGEF1A | 0.77 | 0.82 |
| SLC7A4 | 0.76 | 0.83 |
| SYN1 | 0.75 | 0.81 |
| PRKCG | 0.75 | 0.82 |
| SYN2 | 0.75 | 0.81 |
| SLC30A3 | 0.75 | 0.77 |
| CHRM1 | 0.75 | 0.77 |
| TMEM130 | 0.75 | 0.81 |
| PNMAL2 | 0.74 | 0.77 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| RBMX2 | -0.39 | -0.45 |
| KIAA1949 | -0.38 | -0.26 |
| CSRP2 | -0.37 | -0.43 |
| SLA | -0.37 | -0.02 |
| TUBB2B | -0.37 | -0.36 |
| SH2D2A | -0.37 | -0.41 |
| FABP7 | -0.36 | -0.39 |
| ZNF435 | -0.36 | -0.24 |
| DACT1 | -0.36 | 0.00 |
| MYCN | -0.36 | -0.03 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0005070 | SH3/SH2 adaptor activity | TAS | 9020193 | |
| GO:0005515 | protein binding | IPI | 8681387 |9763511 | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0004715 | non-membrane spanning protein tyrosine kinase activity | IEA | - | |
| GO:0016740 | transferase activity | IEA | - | |
| GO:0042169 | SH2 domain binding | IPI | 8070403 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007172 | signal complex assembly | TAS | 9924018 | |
| GO:0006468 | protein amino acid phosphorylation | IEA | - | |
| GO:0007265 | Ras protein signal transduction | EXP | 11520933 | |
| GO:0007243 | protein kinase cascade | TAS | 9020193 | |
| GO:0044419 | interspecies interaction between organisms | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005829 | cytosol | EXP | 10858437 |11124251 | |
| GO:0005901 | caveola | IDA | 17848177 | |
| GO:0005886 | plasma membrane | EXP | 8845374 |9028946 |11124251 |12506110 |12907686 |15574420 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ABL1 | ABL | JTK7 | bcr/abl | c-ABL | p150 | v-abl | c-abl oncogene 1, receptor tyrosine kinase | c-Abl interacts with Src. This interaction was modeled on a demonstrated interaction between human c-Abl and v-Src from the Rous Sarcoma virus. | BIND | 1383690 |
| ADAM12 | MCMP | MCMPMltna | MLTN | MLTNA | ADAM metallopeptidase domain 12 | - | HPRD | 11127814 |
| ADAM15 | MDC15 | ADAM metallopeptidase domain 15 | - | HPRD | 11741929 |
| ADRB2 | ADRB2R | ADRBR | B2AR | BAR | BETA2AR | adrenergic, beta-2-, receptor, surface | Affinity Capture-Western | BioGRID | 11013230 |
| ADRB3 | BETA3AR | adrenergic, beta-3-, receptor | - | HPRD,BioGRID | 11013230 |
| ADRBK1 | BARK1 | BETA-ARK1 | FLJ16718 | GRK2 | adrenergic, beta, receptor kinase 1 | - | HPRD,BioGRID | 12624098 |
| AFAP1 | AFAP | AFAP-110 | FLJ56849 | actin filament associated protein 1 | - | HPRD,BioGRID | 9655255 |
| AHR | - | aryl hydrocarbon receptor | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 12024042 |
| ANKRD11 | ANCO-1 | LZ16 | T13 | ankyrin repeat domain 11 | - | HPRD | 15184363 |
| ANXA2 | ANX2 | ANX2L4 | CAL1H | LIP2 | LPC2 | LPC2D | P36 | PAP-IV | annexin A2 | - | HPRD,BioGRID | 11106497 |
| AR | AIS | DHTR | HUMARA | KD | NR3C4 | SBMA | SMAX1 | TFM | androgen receptor | Affinity Capture-Western Two-hybrid | BioGRID | 11032808 |15027889 |15466214 |
| ARHGAP1 | CDC42GAP | RHOGAP | RHOGAP1 | p50rhoGAP | Rho GTPase activating protein 1 | - | HPRD,BioGRID | 8253717 |
| ARHGAP17 | DKFZp564A1363 | FLJ37567 | FLJ43368 | MGC87805 | MST066 | MST110 | MSTP038 | MSTP066 | MSTP110 | NADRIN | RICH1 | WBP15 | Rho GTPase activating protein 17 | Reconstituted Complex | BioGRID | 11431473 |
| ARNT | HIF-1beta | HIF1B | HIF1BETA | TANGO | bHLHe2 | aryl hydrocarbon receptor nuclear translocator | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 12024042 |
| ARR3 | ARRX | arrestin 3, retinal (X-arrestin) | - | HPRD,BioGRID | 11877451 |
| ASAP1 | AMAP1 | CENTB4 | DDEF1 | KIAA1249 | PAG2 | PAP | ZG14P | ArfGAP with SH3 domain, ankyrin repeat and PH domain 1 | - | HPRD,BioGRID | 9819391 |
| ASAP2 | AMAP2 | CENTB3 | DDEF2 | FLJ42910 | KIAA0400 | PAG3 | PAP | Pap-alpha | SHAG1 | ArfGAP with SH3 domain, ankyrin repeat and PH domain 2 | Affinity Capture-Western | BioGRID | 10022920 |
| AXL | JTK11 | UFO | AXL receptor tyrosine kinase | - | HPRD,BioGRID | 9178760 |
| BCAR1 | CAS | CAS1 | CASS1 | CRKAS | P130Cas | breast cancer anti-estrogen resistance 1 | c-Src interacts with CAS. | BIND | 9148935 |
| BCAR1 | CAS | CAS1 | CASS1 | CRKAS | P130Cas | breast cancer anti-estrogen resistance 1 | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 10085298 |10487518 |10739664 |11514617 |12397603 |12615911 |
| BCR | ALL | BCR-ABL1 | BCR1 | CML | D22S11 | D22S662 | FLJ16453 | PHL | breakpoint cluster region | BCR interacts with Src. This interaction was modeled on a demonstrated interaction between BCR from an unspecified source and Src from Rous Sarcoma virus. | BIND | 1383690 |
| BMX | ETK | PSCTK2 | PSCTK3 | BMX non-receptor tyrosine kinase | - | HPRD,BioGRID | 10688651 |
| CAV1 | CAV | MSTP085 | VIP21 | caveolin 1, caveolae protein, 22kDa | - | HPRD,BioGRID | 8910575 |
| CAV2 | CAV | MGC12294 | caveolin 2 | Reconstituted Complex | BioGRID | 12091389 |
| CBL | C-CBL | CBL2 | RNF55 | Cas-Br-M (murine) ecotropic retroviral transforming sequence | - | HPRD | 8635998 |
| CBL | C-CBL | CBL2 | RNF55 | Cas-Br-M (murine) ecotropic retroviral transforming sequence | Cbl interacts with Src. This interaction was modeled on a demonstrated interaction between human Cbl and Src from unspecified species. | BIND | 15135048 |
| CD2AP | CMS | DKFZp586H0519 | CD2-associated protein | - | HPRD | 10339567 |
| CD36 | CHDS7 | FAT | GP3B | GP4 | GPIV | PASIV | SCARB3 | CD36 molecule (thrombospondin receptor) | - | HPRD | 7521304 |
| CD44 | CDW44 | CSPG8 | ECMR-III | HCELL | IN | LHR | MC56 | MDU2 | MDU3 | MGC10468 | MIC4 | MUTCH-I | Pgp1 | CD44 molecule (Indian blood group) | Affinity Capture-Western Reconstituted Complex | BioGRID | 11084024 |
| CD46 | MCP | MGC26544 | MIC10 | TLX | TRA2.10 | CD46 molecule, complement regulatory protein | - | HPRD,BioGRID | 10657632 |
| CD59 | 16.3A5 | 1F5 | EJ16 | EJ30 | EL32 | FLJ38134 | FLJ92039 | G344 | HRF-20 | HRF20 | MAC-IP | MACIF | MEM43 | MGC2354 | MIC11 | MIN1 | MIN2 | MIN3 | MIRL | MSK21 | p18-20 | CD59 molecule, complement regulatory protein | - | HPRD | 12219031 |
| CDC37 | P50CDC37 | cell division cycle 37 homolog (S. cerevisiae) | Reconstituted Complex | BioGRID | 11085988 |
| CDCP1 | CD318 | SIMA135 | TRASK | CUB domain containing protein 1 | Src interacts with CDCP1. This interaction was modelled on a demonstrated interaction between Src from an unspecified species and human CDCP1. | BIND | 15851033 |
| CDH1 | Arc-1 | CD324 | CDHE | ECAD | LCAM | UVO | cadherin 1, type 1, E-cadherin (epithelial) | Affinity Capture-Western | BioGRID | 10744074 |
| CEACAM3 | CD66D | CEA | CGM1 | MGC119875 | W264 | W282 | carcinoembryonic antigen-related cell adhesion molecule 3 | - | HPRD | 11708798 |
| CRK | CRKII | v-crk sarcoma virus CT10 oncogene homolog (avian) | Affinity Capture-Western | BioGRID | 12615911 |
| CSF1R | C-FMS | CD115 | CSFR | FIM2 | FMS | colony stimulating factor 1 receptor | Affinity Capture-Western | BioGRID | 7681396 |
| CSK | MGC117393 | c-src tyrosine kinase | - | HPRD,BioGRID | 7513429 |
| CTNND1 | CAS | CTNND | KIAA0384 | P120CAS | P120CTN | p120 | catenin (cadherin-associated protein), delta 1 | Affinity Capture-Western | BioGRID | 12640114 |
| DAB2 | DOC-2 | DOC2 | FLJ26626 | disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila) | - | HPRD,BioGRID | 12473651 |
| DAG1 | 156DAG | A3a | AGRNR | DAG | dystroglycan 1 (dystrophin-associated glycoprotein 1) | Affinity Capture-Western Biochemical Activity Reconstituted Complex | BioGRID | 11724572 |
| DDR2 | MIG20a | NTRKR3 | TKT | TYRO10 | discoidin domain receptor tyrosine kinase 2 | - | HPRD,BioGRID | 11884411 |
| DOCK1 | DOCK180 | ced5 | dedicator of cytokinesis 1 | Affinity Capture-Western | BioGRID | 12615911 |
| DOK1 | MGC117395 | MGC138860 | P62DOK | docking protein 1, 62kDa (downstream of tyrosine kinase 1) | Reconstituted Complex | BioGRID | 11071635 |
| DOK4 | FLJ10488 | docking protein 4 | - | HPRD | 12730241 |
| DOK4 | FLJ10488 | docking protein 4 | IRS5/DOK4 interacts with Src. This interaction was modelled on a demonstrated interaction between human IRS5/DOK4 and hamster Src. | BIND | 12730241 |
| DRD4 | D4DR | dopamine receptor D4 | D4.4 interacts with c-Src. This interaction was modeled on a demonstrated interaction between human D4.4 and chicken c-Src. | BIND | 9843378 |
| DTNA | D18S892E | DRP3 | DTN | FLJ96209 | LVNC1 | dystrobrevin, alpha | - | HPRD | 9701558 |
| E2F4 | E2F-4 | E2F transcription factor 4, p107/p130-binding | E2F4 interacts with SRC enhancer. | BIND | 15782160 |
| EFNB1 | CFND | CFNS | EFL3 | EPLG2 | Elk-L | LERK2 | MGC8782 | ephrin-B1 | - | HPRD,BioGRID | 8878483 |
| EGFR | ERBB | ERBB1 | HER1 | PIG61 | mENA | epidermal growth factor receptor (erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian) | - | HPRD,BioGRID | 10971656 |
| EP300 | KAT3B | p300 | E1A binding protein p300 | Reconstituted Complex | BioGRID | 10973497 |
| EPHB2 | CAPB | DRT | EPHT3 | ERK | Hek5 | MGC87492 | PCBC | Tyro5 | EPH receptor B2 | - | HPRD,BioGRID | 9632142 |
| EPS8 | - | epidermal growth factor receptor pathway substrate 8 | - | HPRD,BioGRID | 10395945 |
| ERBB2 | CD340 | HER-2 | HER-2/neu | HER2 | NEU | NGL | TKR1 | v-erb-b2 erythroblastic leukemia viral oncogene homolog 2, neuro/glioblastoma derived oncogene homolog (avian) | - | HPRD | 7542762 |
| ERBB2 | CD340 | HER-2 | HER-2/neu | HER2 | NEU | NGL | TKR1 | v-erb-b2 erythroblastic leukemia viral oncogene homolog 2, neuro/glioblastoma derived oncogene homolog (avian) | Reconstituted Complex | BioGRID | 11940572 |
| ERBB2 | CD340 | HER-2 | HER-2/neu | HER2 | NEU | NGL | TKR1 | v-erb-b2 erythroblastic leukemia viral oncogene homolog 2, neuro/glioblastoma derived oncogene homolog (avian) | SRC (SRC-1) interacts with the ERBB2 (HER2) promoter. | BIND | 15788656 |
| ESR1 | DKFZp686N23123 | ER | ESR | ESRA | Era | NR3A1 | estrogen receptor 1 | Affinity Capture-Western Reconstituted Complex | BioGRID | 10454579 |11032808 |11564893 |
| ESR2 | ER-BETA | ESR-BETA | ESRB | ESTRB | Erb | NR3A2 | estrogen receptor 2 (ER beta) | Affinity Capture-Western | BioGRID | 11032808 |
| ETS1 | ETS-1 | EWSR2 | FLJ10768 | v-ets erythroblastosis virus E26 oncogene homolog 1 (avian) | SRC (SRC-1) interacts with ETS1. | BIND | 15788656 |
| ETS2 | ETS2IT1 | v-ets erythroblastosis virus E26 oncogene homolog 2 (avian) | SRC (SRC-1) interacts with ETS2. | BIND | 15788656 |
| EVL | RNB6 | Enah/Vasp-like | - | HPRD,BioGRID | 10945997 |
| FARP2 | FIR | FRG | KIAA0793 | PLEKHC3 | FERM, RhoGEF and pleckstrin domain protein 2 | - | HPRD | 12771149 |
| FASLG | APT1LG1 | CD178 | CD95L | FASL | TNFSF6 | Fas ligand (TNF superfamily, member 6) | - | HPRD | 11741599 |
| FOXO1 | FKH1 | FKHR | FOXO1A | forkhead box O1 | - | HPRD,BioGRID | 10973497 |
| FYN | MGC45350 | SLK | SYN | FYN oncogene related to SRC, FGR, YES | Protein-peptide | BioGRID | 9169421 |
| GAB2 | KIAA0571 | GRB2-associated binding protein 2 | - | HPRD,BioGRID | 10391903 |
| GAB3 | - | GRB2-associated binding protein 3 | - | HPRD,BioGRID | 11739737 |
| GFAP | FLJ45472 | glial fibrillary acidic protein | - | HPRD | 11379820 |
| GIT1 | - | G protein-coupled receptor kinase interacting ArfGAP 1 | - | HPRD | 14523024 |
| GJA1 | CX43 | DFNB38 | GJAL | ODDD | gap junction protein, alpha 1, 43kDa | - | HPRD | 9278444 |
| GJA1 | CX43 | DFNB38 | GJAL | ODDD | gap junction protein, alpha 1, 43kDa | Cx43 interacts with v-Src. This interaction was modelled on a demonstrated interaction between rat Cx43 and human v-Src. | BIND | 9278444 |
| GJB1 | CMTX | CMTX1 | CX32 | gap junction protein, beta 1, 32kDa | GJB1 (connexin32) interacts with and prevents the activation of Src. | BIND | 15782139 |
| GNB2L1 | Gnb2-rs1 | H12.3 | HLC-7 | PIG21 | RACK1 | guanine nucleotide binding protein (G protein), beta polypeptide 2-like 1 | - | HPRD,BioGRID | 9584165 |
| GRB10 | GRB-IR | Grb-10 | IRBP | KIAA0207 | MEG1 | RSS | growth factor receptor-bound protein 10 | Biochemical Activity | BioGRID | 10871840 |
| GRB2 | ASH | EGFRBP-GRB2 | Grb3-3 | MST084 | MSTP084 | growth factor receptor-bound protein 2 | - | HPRD,BioGRID | 11964172 |
| GRIN2A | NMDAR2A | NR2A | glutamate receptor, ionotropic, N-methyl D-aspartate 2A | Affinity Capture-Western Reconstituted Complex | BioGRID | 10458595 |12932824 |
| GRIN2A | NMDAR2A | NR2A | glutamate receptor, ionotropic, N-methyl D-aspartate 2A | - | HPRD | 10458595 |10884433 |12932824 |
| GRIN2B | MGC142178 | MGC142180 | NMDAR2B | NR2B | hNR3 | glutamate receptor, ionotropic, N-methyl D-aspartate 2B | - | HPRD | 10884433 |
| GSN | DKFZp313L0718 | gelsolin (amyloidosis, Finnish type) | Affinity Capture-Western | BioGRID | 11577104 |
| HNF1A | HNF1 | LFB1 | MODY3 | TCF1 | HNF1 homeobox A | - | HPRD,BioGRID | 10777539 |
| HNRNPK | CSBP | FLJ41122 | HNRPK | TUNP | heterogeneous nuclear ribonucleoprotein K | Src interacts with hnRNPK. This interaction was modeled on a demonstrated interaction between Src from an unspecified source and human hnRNPK. | BIND | 8810341 |
| HSP90AA1 | FLJ31884 | HSP86 | HSP89A | HSP90A | HSP90N | HSPC1 | HSPCA | HSPCAL1 | HSPCAL4 | HSPN | Hsp89 | Hsp90 | LAP2 | heat shock protein 90kDa alpha (cytosolic), class A member 1 | - | HPRD,BioGRID | 1310678 |
| IL6R | CD126 | IL-6R-1 | IL-6R-alpha | IL6RA | MGC104991 | interleukin 6 receptor | - | HPRD | 10462375 |
| ITK | EMT | LYK | MGC126257 | MGC126258 | PSCTK2 | IL2-inducible T-cell kinase | Reconstituted Complex | BioGRID | 10636929 |
| KCNQ5 | Kv7.5 | potassium voltage-gated channel, KQT-like subfamily, member 5 | - | HPRD | 15304482 |
| KHDRBS1 | FLJ34027 | Sam68 | p62 | KH domain containing, RNA binding, signal transduction associated 1 | - | HPRD,BioGRID | 1545818 |8766817 |10467411 |
| KHDRBS1 | FLJ34027 | Sam68 | p62 | KH domain containing, RNA binding, signal transduction associated 1 | c-Src SH3 domain interacts with Sam68 proline-rich region. This interaction was modelled on a demonstrated interaction between c-Src from an unspecified source and human Sam68. | BIND | 7537265 |
| KIFAP3 | FLJ22818 | KAP3 | SMAP | Smg-GDS | dJ190I16.1 | kinesin-associated protein 3 | - | HPRD,BioGRID | 8900189 |
| LRP1 | A2MR | APOER | APR | CD91 | FLJ16451 | IGFBP3R | LRP | MGC88725 | TGFBR5 | low density lipoprotein-related protein 1 (alpha-2-macroglobulin receptor) | Biochemical Activity | BioGRID | 11854294 |
| LYN | FLJ26625 | JTK8 | v-yes-1 Yamaguchi sarcoma viral related oncogene homolog | Protein-peptide | BioGRID | 9169421 |
| MAP2 | DKFZp686I2148 | MAP2A | MAP2B | MAP2C | microtubule-associated protein 2 | Reconstituted Complex | BioGRID | 10781592 |
| MAPK15 | ERK7 | ERK8 | mitogen-activated protein kinase 15 | - | HPRD | 11875070 |
| MAPK8IP3 | DKFZp762N1113 | FLJ00027 | JIP3 | JSAP1 | KIAA1066 | SYD2 | mitogen-activated protein kinase 8 interacting protein 3 | Biochemical Activity | BioGRID | 12226752 |
| MATK | CHK | CTK | DKFZp434N1212 | HHYLTK | HYL | HYLTK | Lsk | MGC1708 | MGC2101 | megakaryocyte-associated tyrosine kinase | - | HPRD,BioGRID | 7530249 |
| MET | AUTS9 | HGFR | RCCP2 | c-Met | met proto-oncogene (hepatocyte growth factor receptor) | - | HPRD,BioGRID | 9837958 |
| MICAL1 | DKFZp434B1517 | FLJ11937 | FLJ21739 | MICAL | MICAL-1 | NICAL | microtubule associated monoxygenase, calponin and LIM domain containing 1 | Reconstituted Complex | BioGRID | 11827972 |
| MPZL1 | FLJ21047 | PZR | PZR1b | PZRa | PZRb | myelin protein zero-like 1 | - | HPRD,BioGRID | 11751924 |
| MST1R | CD136 | CDw136 | PTK8 | RON | macrophage stimulating 1 receptor (c-met-related tyrosine kinase) | - | HPRD | 8918464 |
| MUC1 | CD227 | EMA | H23AG | MAM6 | PEM | PEMT | PUM | mucin 1, cell surface associated | MUC1 interacts with c-Src. | BIND | 11483589 |
| MUC1 | CD227 | EMA | H23AG | MAM6 | PEM | PEMT | PUM | mucin 1, cell surface associated | - | HPRD,BioGRID | 11152665 |11483589 |
| MYLK | DKFZp686I10125 | FLJ12216 | KRP | MLCK | MLCK1 | MLCK108 | MLCK210 | MSTP083 | MYLK1 | smMLCK | myosin light chain kinase | - | HPRD,BioGRID | 10362724 |
| NCOA6 | AIB3 | ANTP | ASC2 | HOX1.1 | HOXA7 | KIAA0181 | NRC | PRIP | RAP250 | TRBP | nuclear receptor coactivator 6 | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 10567404 |10847592 |11158331 |12482968 |
| ND2 | MTND2 | NADH dehydrogenase, subunit 2 (complex I) | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 15069201 |
| NPHS1 | CNF | NPHN | nephrosis 1, congenital, Finnish type (nephrin) | - | HPRD | 12846735 |
| PDE4D | DPDE3 | HSPDE4D | PDE4DN2 | STRK1 | phosphodiesterase 4D, cAMP-specific (phosphodiesterase E3 dunce homolog, Drosophila) | PDE4D4 interacts with Src SH3 domain. | BIND | 10571082 |
| PDE6G | DKFZp686C0587 | MGC125749 | PDEG | phosphodiesterase 6G, cGMP-specific, rod, gamma | - | HPRD,BioGRID | 12624098 |
| PDGFRB | CD140B | JTK12 | PDGF-R-beta | PDGFR | PDGFR1 | platelet-derived growth factor receptor, beta polypeptide | PDGFR interacts with Src. This interaction was modelled on a demonstrated interaction between rat PDGFR and Src from an unknown species. | BIND | 2173144 |
| PECAM1 | CD31 | PECAM-1 | platelet/endothelial cell adhesion molecule | - | HPRD,BioGRID | 10858437 |
| PELP1 | HMX3 | MNAR | P160 | proline, glutamate and leucine rich protein 1 | Affinity Capture-Western | BioGRID | 15466214 |
| PIK3R1 | GRB1 | p85 | p85-ALPHA | phosphoinositide-3-kinase, regulatory subunit 1 (alpha) | - | HPRD | 12210743 |
| PIK3R1 | GRB1 | p85 | p85-ALPHA | phosphoinositide-3-kinase, regulatory subunit 1 (alpha) | Affinity Capture-Western | BioGRID | 10487518 |
| PKD1 | PBP | polycystic kidney disease 1 (autosomal dominant) | - | HPRD | 11113628 |
| PLCG1 | PLC-II | PLC1 | PLC148 | PLCgamma1 | phospholipase C, gamma 1 | - | HPRD,BioGRID | 7510703 |
| PLD1 | - | phospholipase D1, phosphatidylcholine-specific | Biochemical Activity | BioGRID | 12697812 |
| PLD2 | - | phospholipase D2 | Affinity Capture-Western Biochemical Activity Reconstituted Complex | BioGRID | 12697812 |
| PLSCR1 | MMTRA1B | phospholipid scramblase 1 | Biochemical Activity | BioGRID | 12871937 |
| PRKCE | MGC125656 | MGC125657 | PKCE | nPKC-epsilon | protein kinase C, epsilon | - | HPRD | 11834516 |
| PRKCZ | PKC-ZETA | PKC2 | protein kinase C, zeta | - | HPRD,BioGRID | 10527887 |
| PTK2 | FADK | FAK | FAK1 | pp125FAK | PTK2 protein tyrosine kinase 2 | c-Src interacts with FAK. This interaction was modeled on a demonstrated interaction between human c-Src and mouse FAK. | BIND | 9148935 |
| PTK2 | FADK | FAK | FAK1 | pp125FAK | PTK2 protein tyrosine kinase 2 | Affinity Capture-Western Reconstituted Complex | BioGRID | 10085298 |11980671 |12387730 |12558988 |12615911 |
| PTK2 | FADK | FAK | FAK1 | pp125FAK | PTK2 protein tyrosine kinase 2 | - | HPRD | 7529876 |8054685 |10931187 |
| PTK2 | FADK | FAK | FAK1 | pp125FAK | PTK2 protein tyrosine kinase 2 | - | HPRD | 7529876 |8054685 |
| PTK2 | FADK | FAK | FAK1 | pp125FAK | PTK2 protein tyrosine kinase 2 | Src interacts with tyrosine autophosphorylated FAK. This interaction was modeled on a demonstrated interaction between human Src and avian FAK. | BIND | 7515480 |
| PTK2B | CADTK | CAKB | FADK2 | FAK2 | FRNK | PKB | PTK | PYK2 | RAFTK | PTK2B protein tyrosine kinase 2 beta | - | HPRD | 8849729 |10777553 |
| PTK2B | CADTK | CAKB | FADK2 | FAK2 | FRNK | PKB | PTK | PYK2 | RAFTK | PTK2B protein tyrosine kinase 2 beta | Affinity Capture-Western Reconstituted Complex | BioGRID | 8849729 |10521452 |10777553 |
| PTPN2 | PTPT | TC-PTP | TCELLPTP | TCPTP | protein tyrosine phosphatase, non-receptor type 2 | PTPN2 (TCPTP) interacts with Src. This interaction was modelled on a demonstrated interaction between human PTPN2 and chicken Src. | BIND | 15696169 |
| PTPN21 | PTPD1 | PTPRL10 | protein tyrosine phosphatase, non-receptor type 21 | - | HPRD,BioGRID | 7519780 |
| PTPRA | HEPTP | HLPR | HPTPA | HPTPalpha | LRP | PTPA | PTPRL2 | R-PTP-alpha | RPTPA | protein tyrosine phosphatase, receptor type, A | Affinity Capture-Western | BioGRID | 11923305 |
| PXN | FLJ16691 | paxillin | Reconstituted Complex | BioGRID | 10085298 |
| PXN | FLJ16691 | paxillin | - | HPRD | 7534286 |
| RAF1 | CRAF | NS5 | Raf-1 | c-Raf | v-raf-1 murine leukemia viral oncogene homolog 1 | - | HPRD,BioGRID | 7517401 |
| RARA | NR1B1 | RAR | retinoic acid receptor, alpha | Reconstituted Complex | BioGRID | 10454579 |
| RASA1 | CM-AVM | CMAVM | DKFZp434N071 | GAP | PKWS | RASA | RASGAP | p120GAP | p120RASGAP | RAS p21 protein activator (GTPase activating protein) 1 | - | HPRD,BioGRID | 1717825 |
| RET | CDHF12 | HSCR1 | MEN2A | MEN2B | MTC1 | PTC | RET-ELE1 | RET51 | ret proto-oncogene | - | HPRD,BioGRID | 10070972 |
| RGS16 | A28-RGS14 | A28-RGS14P | RGS-R | regulator of G-protein signaling 16 | SRC phosphorylates RGS16. This interaction was modeled on a demonstrated interaction between SRC from an unspecified species and human RSG16. | BIND | 12588871 |
| RICS | GC-GAP | GRIT | KIAA0712 | MGC1892 | p200RhoGAP | p250GAP | Rho GTPase-activating protein | - | HPRD,BioGRID | 12454018 |
| RICS | GC-GAP | GRIT | KIAA0712 | MGC1892 | p200RhoGAP | p250GAP | Rho GTPase-activating protein | Src interacts with GC-GAP. This interaction was modelled on a demonstrated interaction between Src from an unspecified species and human GC-GAP. | BIND | 12819203 |
| RPL10 | DKFZp686J1851 | DXS648 | DXS648E | FLJ23544 | FLJ27072 | NOV | QM | ribosomal protein L10 | Reconstituted Complex | BioGRID | 12138090 |
| RPS6KB1 | PS6K | S6K | S6K1 | STK14A | p70(S6K)-alpha | p70-S6K | p70-alpha | ribosomal protein S6 kinase, 70kDa, polypeptide 1 | Biochemical Activity | BioGRID | 16640565 |
| RPS6KB2 | KLS | P70-beta | P70-beta-1 | P70-beta-2 | S6K-beta2 | S6K2 | SRK | STK14B | p70(S6K)-beta | p70S6Kb | ribosomal protein S6 kinase, 70kDa, polypeptide 2 | Biochemical Activity | BioGRID | 16640565 |
| RXRA | FLJ00280 | FLJ00318 | FLJ16020 | FLJ16733 | MGC102720 | NR2B1 | retinoid X receptor, alpha | Reconstituted Complex | BioGRID | 10517671 |
| SH3BP1 | - | SH3-domain binding protein 1 | - | HPRD,BioGRID | 1379745 |
| SH3PXD2A | FISH | SH3MD1 | SH3 and PX domains 2A | - | HPRD | 9687503 |
| SHB | RP11-3J10.8 | bA3J10.2 | Src homology 2 domain containing adaptor protein B | Shb interacts with Src. This interaction was modelled on a demonstrated interaction between human Shb and chicken Src. | BIND | 7537362 |
| SHB | RP11-3J10.8 | bA3J10.2 | Src homology 2 domain containing adaptor protein B | - | HPRD,BioGRID | 7537362 |
| SHC1 | FLJ26504 | SHC | SHCA | SHC (Src homology 2 domain containing) transforming protein 1 | Src interacts with Shc. This interaction was modelled on demonstrated interactions between human Src and rat Shc, and between Src from unspecified species and human Shc. | BIND | 7515480 |8939605 |
| SKAP1 | SCAP1 | SKAP55 | src kinase associated phosphoprotein 1 | - | HPRD,BioGRID | 9195899 |
| SLC9A2 | NHE2 | solute carrier family 9 (sodium/hydrogen exchanger), member 2 | - | HPRD,BioGRID | 10187839 |
| SMARCB1 | BAF47 | INI1 | RDT | SNF5 | SNF5L1 | Sfh1p | Snr1 | hSNFS | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily b, member 1 | - | HPRD | 12145209 |
| SMARCE1 | BAF57 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily e, member 1 | - | HPRD | 12145209 |
| SNURF | - | SNRPN upstream reading frame | Phenotypic Enhancement | BioGRID | 11696545 |
| SRC | ASV | SRC1 | c-SRC | p60-Src | v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (avian) | Tyrosine phophorylated c-Src interacts with Src SH2 domain. | BIND | 7683128 |
| SRC | ASV | SRC1 | c-SRC | p60-Src | v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (avian) | - | HPRD | 9174343 |
| SRF | MCM1 | serum response factor (c-fos serum response element-binding transcription factor) | - | HPRD,BioGRID | 9786846 |
| STAT1 | DKFZp686B04100 | ISGF-3 | STAT91 | signal transducer and activator of transcription 1, 91kDa | - | HPRD,BioGRID | 9344858 |
| STAT3 | APRF | FLJ20882 | HIES | MGC16063 | signal transducer and activator of transcription 3 (acute-phase response factor) | - | HPRD,BioGRID | 8657134 |
| STAT5A | MGF | STAT5 | signal transducer and activator of transcription 5A | - | HPRD | 10358079 |
| SYK | DKFZp313N1010 | FLJ25043 | FLJ37489 | spleen tyrosine kinase | - | HPRD,BioGRID | 7513017 |
| SYN1 | SYN1a | SYN1b | SYNI | synapsin I | - | HPRD,BioGRID | 10899172 |
| THRB | ERBA-BETA | ERBA2 | GRTH | MGC126109 | MGC126110 | NR1A2 | PRTH | THR1 | THRB1 | THRB2 | thyroid hormone receptor, beta (erythroblastic leukemia viral (v-erb-a) oncogene homolog 2, avian) | Reconstituted Complex | BioGRID | 10454579 |
| TIAM1 | FLJ36302 | T-cell lymphoma invasion and metastasis 1 | - | HPRD | 12810717 |
| TNFSF11 | CD254 | ODF | OPGL | OPTB2 | RANKL | TRANCE | hRANKL2 | sOdf | tumor necrosis factor (ligand) superfamily, member 11 | Affinity Capture-Western | BioGRID | 10635328 |
| TRAF1 | EBI6 | MGC:10353 | TNF receptor-associated factor 1 | Affinity Capture-Western | BioGRID | 10635328 |
| TRAF3 | CAP-1 | CD40bp | CRAF1 | LAP1 | TNF receptor-associated factor 3 | Affinity Capture-Western | BioGRID | 10635328 |
| TRAF6 | MGC:3310 | RNF85 | TNF receptor-associated factor 6 | - | HPRD,BioGRID | 10635328 |
| TRIP4 | HsT17391 | thyroid hormone receptor interactor 4 | Affinity Capture-Western | BioGRID | 10454579 |
| TRIP6 | MGC10556 | MGC10558 | MGC29959 | MGC3837 | MGC4423 | OIP1 | ZRP-1 | thyroid hormone receptor interactor 6 | - | HPRD,BioGRID | 14688263 |
| TRPC6 | FLJ11098 | FLJ14863 | FSGS2 | TRP6 | transient receptor potential cation channel, subfamily C, member 6 | - | HPRD,BioGRID | 14761972 |
| TRPV4 | OTRPC4 | TRP12 | VR-OAC | VRL-2 | VRL2 | VROAC | transient receptor potential cation channel, subfamily V, member 4 | - | HPRD,BioGRID | 12538589 |
| TUB | rd5 | tubby homolog (mouse) | - | HPRD | 10455176 |
| TYRO3 | BYK | Brt | Dtk | FLJ16467 | RSE | Sky | Tif | TYRO3 protein tyrosine kinase | - | HPRD,BioGRID | 7537495 |
| WAS | IMD2 | THC | WASP | Wiskott-Aldrich syndrome (eczema-thrombocytopenia) | - | HPRD,BioGRID | 8805332 |
| WBP11 | DKFZp779M1063 | NPWBP | SIPP1 | WW domain binding protein 11 | - | HPRD | 11375989 |
| YTHDC1 | KIAA1966 | YT521 | YT521-B | YTH domain containing 1 | - | HPRD | 15175272 |
| YWHAB | GW128 | HS1 | KCIP-1 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide | - | HPRD | 8702721 |
| YWHAE | 14-3-3E | FLJ45465 | KCIP-1 | MDCR | MDS | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon polypeptide | - | HPRD | 8702721 |
| YWHAG | 14-3-3GAMMA | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide | - | HPRD | 8702721 |
| YWHAH | YWHA1 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta polypeptide | - | HPRD | 8702721 |
Section V. Pathway annotation
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-137 | 1620 | 1626 | 1A | hsa-miR-137 | UAUUGCUUAAGAAUACGCGUAG |
| miR-141/200a | 1867 | 1873 | m8 | hsa-miR-141 | UAACACUGUCUGGUAAAGAUGG |
| hsa-miR-200a | UAACACUGUCUGGUAACGAUGU | ||||
| miR-149 | 1651 | 1657 | m8 | hsa-miR-149brain | UCUGGCUCCGUGUCUUCACUCC |
| miR-153 | 1891 | 1897 | m8 | hsa-miR-153 | UUGCAUAGUCACAAAAGUGA |
| miR-203.1 | 1594 | 1601 | 1A,m8 | hsa-miR-203 | UGAAAUGUUUAGGACCACUAG |
| hsa-miR-203 | UGAAAUGUUUAGGACCACUAG | ||||
| miR-378 | 764 | 770 | 1A | hsa-miR-378 | CUCCUGACUCCAGGUCCUGUGU |
| miR-9 | 419 | 425 | 1A | hsa-miR-9SZ | UCUUUGGUUAUCUAGCUGUAUGA |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.