Gene Page: SRF
Summary ?
| GeneID | 6722 |
| Symbol | SRF |
| Synonyms | MCM1 |
| Description | serum response factor |
| Reference | MIM:600589|HGNC:HGNC:11291|Ensembl:ENSG00000112658|HPRD:02788|Vega:OTTHUMG00000014722 |
| Gene type | protein-coding |
| Map location | 6p21.1 |
| Pascal p-value | 3.242E-5 |
| Sherlock p-value | 0.286 |
| Fetal beta | 0.645 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Nishioka_2013 | Genome-wide DNA methylation analysis | The authors investigated the methylation profiles of DNA in peripheral blood cells from 18 patients with first-episode schizophrenia (FESZ) and from 15 normal controls. | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| GSMA_IIE | Genome scan meta-analysis (European-ancestry samples) | Psr: 0.04433 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg16041611 | 6 | 43139680 | SRF | -0.087 | 0.51 | DMG:Nishioka_2013 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs1578978 | chr8 | 118196802 | SRF | 6722 | 0.02 | trans |
Section II. Transcriptome annotation
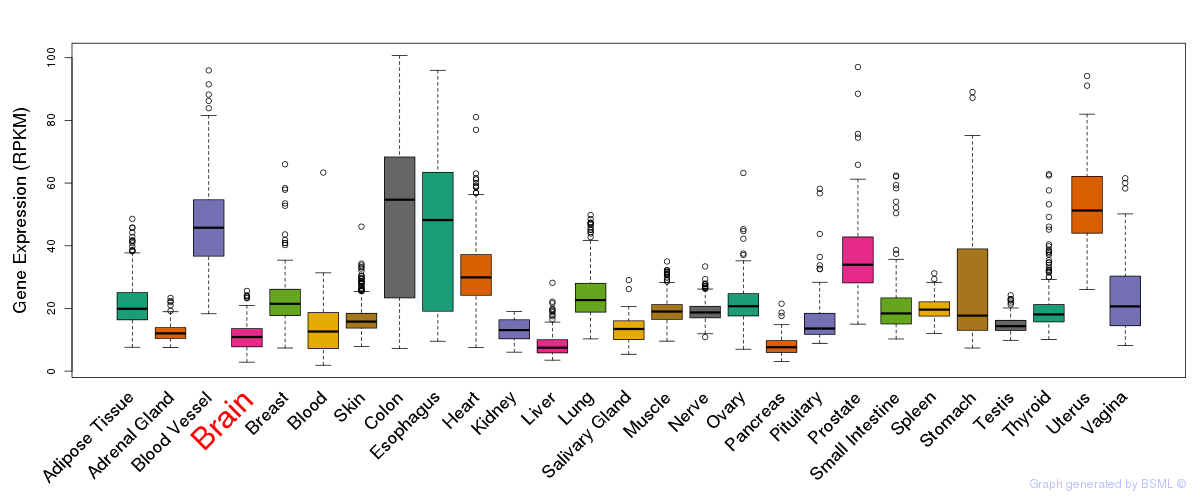
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003700 | transcription factor activity | IEA | - | |
| GO:0003702 | RNA polymerase II transcription factor activity | TAS | 3203386 | |
| GO:0008134 | transcription factor binding | IPI | 16054032 | |
| GO:0043565 | sequence-specific DNA binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0001829 | trophectodermal cell differentiation | IDA | 17576768 | |
| GO:0001947 | heart looping | IEA | - | |
| GO:0006355 | regulation of transcription, DNA-dependent | IEA | - | |
| GO:0006350 | transcription | IEA | - | |
| GO:0007165 | signal transduction | TAS | 3203386 | |
| GO:0007507 | heart development | IEA | - | |
| GO:0007275 | multicellular organismal development | IEA | - | |
| GO:0045944 | positive regulation of transcription from RNA polymerase II promoter | IDA | 17576768 | |
| GO:0045597 | positive regulation of cell differentiation | IDA | 17576768 | |
| GO:0046716 | muscle maintenance | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005634 | nucleus | TAS | 3203386 | |
| GO:0005730 | nucleolus | IDA | 18029348 | |
| GO:0005737 | cytoplasm | IDA | 18029348 | |
| GO:0005783 | endoplasmic reticulum | IDA | 18029348 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ASCC1 | ASC1p50 | CGI-18 | RP11-150D20.4 | activating signal cointegrator 1 complex subunit 1 | Reconstituted Complex Two-hybrid | BioGRID | 12077347 |
| ASCC2 | ASC1p100 | activating signal cointegrator 1 complex subunit 2 | Reconstituted Complex Two-hybrid | BioGRID | 12077347 |
| ASCC3 | ASC1p200 | DJ467N11.1 | HELIC1 | MGC26074 | RNAH | dJ121G13.4 | activating signal cointegrator 1 complex subunit 3 | Reconstituted Complex Two-hybrid | BioGRID | 12077347 |
| ATF6 | ATF6A | activating transcription factor 6 | - | HPRD,BioGRID | 9271374 |
| BARX2 | MGC133368 | MGC133369 | BARX homeobox 2 | - | HPRD | 11278942 |
| CD63 | LAMP-3 | ME491 | MLA1 | OMA81H | TSPAN30 | CD63 molecule | Two-hybrid | BioGRID | 9271374 |
| CEBPB | C/EBP-beta | CRP2 | IL6DBP | LAP | MGC32080 | NF-IL6 | TCF5 | CCAAT/enhancer binding protein (C/EBP), beta | - | HPRD | 11500490 |
| CEBPB | C/EBP-beta | CRP2 | IL6DBP | LAP | MGC32080 | NF-IL6 | TCF5 | CCAAT/enhancer binding protein (C/EBP), beta | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 9032301 |10318842 |
| CREBBP | CBP | KAT3A | RSTS | CREB binding protein | Affinity Capture-Western | BioGRID | 12622724 |
| CRIP2 | CRIP | CRP2 | ESP1 | cysteine-rich protein 2 | - | HPRD | 12530967 |
| ELK1 | - | ELK1, member of ETS oncogene family | - | HPRD,BioGRID | 9010223 |
| ELK4 | SAP1 | ELK4, ETS-domain protein (SRF accessory protein 1) | - | HPRD | 11406578 |11846562 |
| ELK4 | SAP1 | ELK4, ETS-domain protein (SRF accessory protein 1) | - | HPRD,BioGRID | 11406578 |
| FHL1 | FHL1B | FLH1A | KYO-T | MGC111107 | SLIM1 | XMPMA | bA535K18.1 | four and a half LIM domains 1 | SRF interacts with FHL1. This interaction was modeled on a demonstrated interaction between human SRF and FHL1 from an unspecified species. | BIND | 15610731 |
| FHL2 | AAG11 | DRAL | SLIM3 | four and a half LIM domains 2 | SRF interacts with FHL2. | BIND | 15610731 |
| FHL3 | MGC19547 | MGC23614 | MGC8696 | SLIM2 | four and a half LIM domains 3 | SRF interacts with FHL3. This interaction was modeled on a demonstrated interaction between human SRF and FHL3 from an unspecified species. | BIND | 15610731 |
| FHL5 | ACT | FLJ33049 | KIAA0776 | RP3-393D12.2 | dJ393D12.2 | four and a half LIM domains 5 | SRF interacts with ACT. This interaction was modeled on a demonstrated interaction between human SRF and ACT from an unspecified species. | BIND | 15610731 |
| FLI1 | EWSR2 | SIC-1 | Friend leukemia virus integration 1 | - | HPRD,BioGRID | 9010223 |10606656 |
| FOS | AP-1 | C-FOS | v-fos FBJ murine osteosarcoma viral oncogene homolog | SRF interacts with the c-fos promoter CArG box. This interaction was modeled on a demonstrated interaction between human SRF and c-fos promoter CArG box from an unspecified species. | BIND | 15610731 |
| FOSL1 | FRA | FRA1 | fra-1 | FOS-like antigen 1 | SRF interacts with the region of the FRA-1 promoter containing the SRE and ATF sites. | BIND | 15806162 |
| GATA4 | MGC126629 | GATA binding protein 4 | - | HPRD,BioGRID | 11158291 |
| GTF2F1 | BTF4 | RAP74 | TF2F1 | TFIIF | general transcription factor IIF, polypeptide 1, 74kDa | - | HPRD,BioGRID | 7854423 |8106390 |
| GTF2I | BAP-135 | BAP135 | BTKAP1 | DIWS | FLJ38776 | FLJ56355 | IB291 | SPIN | TFII-I | WBS | WBSCR6 | general transcription factor II, i | - | HPRD,BioGRID | 9334314 |
| HOPX | Cameo | HOP | LAGY | MGC20820 | NECC1 | OB1 | SMAP31 | Toto | HOP homeobox | - | HPRD | 12297045 |
| MKL1 | BSAC | MAL | MRTF-A | megakaryoblastic leukemia (translocation) 1 | - | HPRD,BioGRID | 12397177|14565952 |
| MKL1 | BSAC | MAL | MRTF-A | megakaryoblastic leukemia (translocation) 1 | - | HPRD | 14565952 |
| MKL2 | DKFZp686J1745 | FLJ31823 | FLJ45623 | MRTF-B | NPD001 | MKL/myocardin-like 2 | - | HPRD,BioGRID | 12397177 |14565952 |
| MYOCD | MYCD | myocardin | - | HPRD,BioGRID | 11439182 |12397177 |15014501 |
| MYOD1 | MYF3 | MYOD | PUM | bHLHc1 | myogenic differentiation 1 | - | HPRD | 8617811 |
| MYOG | MYF4 | MYOGENIN | bHLHc3 | myogenin (myogenic factor 4) | - | HPRD,BioGRID | 8617811 |
| NCOA1 | F-SRC-1 | KAT13A | MGC129719 | MGC129720 | NCoA-1 | RIP160 | SRC-1 | SRC1 | bHLHe42 | nuclear receptor coactivator 1 | - | HPRD | 9786846 |
| NCOA6 | AIB3 | ANTP | ASC2 | HOX1.1 | HOXA7 | KIAA0181 | NRC | PRIP | RAP250 | TRBP | nuclear receptor coactivator 6 | Reconstituted Complex | BioGRID | 10847592 |
| NCOR2 | CTG26 | SMRT | SMRTE | SMRTE-tau | TNRC14 | TRAC-1 | TRAC1 | nuclear receptor co-repressor 2 | - | HPRD,BioGRID | 10777532 |
| NFYA | CBF-A | CBF-B | FLJ11236 | HAP2 | NF-YA | nuclear transcription factor Y, alpha | NF-YA interacts with SRF. This interaction was modeled on a demonstrated interaction between NF-YA from an unspecified species and SRF from human. | BIND | 10571058 |
| NFYA | CBF-A | CBF-B | FLJ11236 | HAP2 | NF-YA | nuclear transcription factor Y, alpha | Reconstituted Complex Two-hybrid | BioGRID | 10571058 |
| NKX2-3 | CSX3 | NK2.3 | NKX2.3 | NKX2C | NKX4-3 | NK2 transcription factor related, locus 3 (Drosophila) | - | HPRD | 11457859 |
| NKX2-5 | CHNG5 | CSX | CSX1 | NKX2.5 | NKX2E | NKX4-1 | NK2 transcription factor related, locus 5 (Drosophila) | - | HPRD | 8887666 |
| NKX3-1 | BAPX2 | NKX3 | NKX3.1 | NKX3A | NK3 homeobox 1 | - | HPRD,BioGRID | 10993896 |
| PML | MYL | PP8675 | RNF71 | TRIM19 | promyelocytic leukemia | - | HPRD,BioGRID | 12622724 |
| PRRX1 | PHOX1 | PMX1 | PRX1 | paired related homeobox 1 | - | HPRD | 9334314 |
| PYCARD | ASC | CARD5 | MGC10332 | TMS | TMS-1 | TMS1 | PYD and CARD domain containing | - | HPRD | 10847592 |
| RARG | NR1B3 | RARC | retinoic acid receptor, gamma | Reconstituted Complex | BioGRID | 11641790 |
| REL | C-Rel | v-rel reticuloendotheliosis viral oncogene homolog (avian) | Reconstituted Complex | BioGRID | 8670842 |
| RELA | MGC131774 | NFKB3 | p65 | v-rel reticuloendotheliosis viral oncogene homolog A (avian) | Reconstituted Complex | BioGRID | 8670842 |
| RXRA | FLJ00280 | FLJ00318 | FLJ16020 | FLJ16733 | MGC102720 | NR2B1 | retinoid X receptor, alpha | - | HPRD | 11641790 |
| RXRG | NR2B3 | RXRC | retinoid X receptor, gamma | - | HPRD,BioGRID | 11641790 |
| SP1 | - | Sp1 transcription factor | Reconstituted Complex | BioGRID | 10082523 |
| SRC | ASV | SRC1 | c-SRC | p60-Src | v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (avian) | - | HPRD,BioGRID | 9786846 |
| SRF | MCM1 | serum response factor (c-fos serum response element-binding transcription factor) | - | HPRD | 11406578 |
| SSRP1 | FACT | FACT80 | T160 | structure specific recognition protein 1 | - | HPRD,BioGRID | 10336466 |
| TEAD1 | AA | REF1 | TCF13 | TEF-1 | TEA domain family member 1 (SV40 transcriptional enhancer factor) | - | HPRD,BioGRID | 11136726 |12061776 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG MAPK SIGNALING PATHWAY | 267 | 205 | All SZGR 2.0 genes in this pathway |
| BIOCARTA EGF PATHWAY | 31 | 25 | All SZGR 2.0 genes in this pathway |
| BIOCARTA GH PATHWAY | 28 | 22 | All SZGR 2.0 genes in this pathway |
| BIOCARTA IGF1 PATHWAY | 21 | 16 | All SZGR 2.0 genes in this pathway |
| BIOCARTA IL6 PATHWAY | 22 | 16 | All SZGR 2.0 genes in this pathway |
| BIOCARTA INSULIN PATHWAY | 22 | 16 | All SZGR 2.0 genes in this pathway |
| BIOCARTA PDGF PATHWAY | 32 | 25 | All SZGR 2.0 genes in this pathway |
| BIOCARTA MAL PATHWAY | 19 | 17 | All SZGR 2.0 genes in this pathway |
| ST P38 MAPK PATHWAY | 37 | 28 | All SZGR 2.0 genes in this pathway |
| ST GA13 PATHWAY | 37 | 32 | All SZGR 2.0 genes in this pathway |
| PID RHOA PATHWAY | 45 | 33 | All SZGR 2.0 genes in this pathway |
| PID HDAC CLASSII PATHWAY | 34 | 27 | All SZGR 2.0 genes in this pathway |
| PID AR PATHWAY | 61 | 46 | All SZGR 2.0 genes in this pathway |
| PID ERBB1 DOWNSTREAM PATHWAY | 105 | 78 | All SZGR 2.0 genes in this pathway |
| PID PDGFRB PATHWAY | 129 | 103 | All SZGR 2.0 genes in this pathway |
| PID P38 MK2 PATHWAY | 21 | 19 | All SZGR 2.0 genes in this pathway |
| PID PDGFRA PATHWAY | 22 | 18 | All SZGR 2.0 genes in this pathway |
| PID MAPK TRK PATHWAY | 34 | 31 | All SZGR 2.0 genes in this pathway |
| LIU SOX4 TARGETS DN | 309 | 191 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 1 UP | 380 | 236 | All SZGR 2.0 genes in this pathway |
| NAGASHIMA NRG1 SIGNALING UP | 176 | 123 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS UP | 214 | 155 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE DN | 485 | 334 | All SZGR 2.0 genes in this pathway |
| YEMELYANOV GR TARGETS DN | 10 | 8 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN UP | 1142 | 669 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN UP | 612 | 367 | All SZGR 2.0 genes in this pathway |
| DARWICHE SKIN TUMOR PROMOTER DN | 185 | 115 | All SZGR 2.0 genes in this pathway |
| DARWICHE PAPILLOMA RISK LOW DN | 165 | 107 | All SZGR 2.0 genes in this pathway |
| DARWICHE PAPILLOMA RISK HIGH DN | 180 | 110 | All SZGR 2.0 genes in this pathway |
| DARWICHE SQUAMOUS CELL CARCINOMA DN | 181 | 107 | All SZGR 2.0 genes in this pathway |
| MARTIN INTERACT WITH HDAC | 44 | 31 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN UP | 479 | 299 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS DN | 1024 | 594 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS DN | 637 | 377 | All SZGR 2.0 genes in this pathway |
| BENPORATH CYCLING GENES | 648 | 385 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES CORE NINE CORRELATED | 100 | 68 | All SZGR 2.0 genes in this pathway |
| COLLIS PRKDC SUBSTRATES | 20 | 15 | All SZGR 2.0 genes in this pathway |
| MAGRANGEAS MULTIPLE MYELOMA IGG VS IGA UP | 22 | 12 | All SZGR 2.0 genes in this pathway |
| FLECHNER PBL KIDNEY TRANSPLANT REJECTED VS OK UP | 63 | 48 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS EARLY PROGENITOR | 532 | 309 | All SZGR 2.0 genes in this pathway |
| WEIGEL OXIDATIVE STRESS BY TBH AND H2O2 | 36 | 24 | All SZGR 2.0 genes in this pathway |
| GENTILE UV RESPONSE CLUSTER D5 | 39 | 26 | All SZGR 2.0 genes in this pathway |
| ZHU CMV ALL UP | 120 | 89 | All SZGR 2.0 genes in this pathway |
| GENTILE UV HIGH DOSE DN | 312 | 203 | All SZGR 2.0 genes in this pathway |
| ZHU CMV 24 HR UP | 93 | 65 | All SZGR 2.0 genes in this pathway |
| DOUGLAS BMI1 TARGETS UP | 566 | 371 | All SZGR 2.0 genes in this pathway |
| OUYANG PROSTATE CANCER PROGRESSION DN | 21 | 16 | All SZGR 2.0 genes in this pathway |
| ACEVEDO NORMAL TISSUE ADJACENT TO LIVER TUMOR DN | 354 | 216 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER DN | 540 | 340 | All SZGR 2.0 genes in this pathway |
| ACEVEDO METHYLATED IN LIVER CANCER DN | 940 | 425 | All SZGR 2.0 genes in this pathway |
| QI PLASMACYTOMA UP | 259 | 185 | All SZGR 2.0 genes in this pathway |
| PARK APL PATHOGENESIS DN | 50 | 35 | All SZGR 2.0 genes in this pathway |
| HOFFMANN LARGE TO SMALL PRE BII LYMPHOCYTE DN | 72 | 47 | All SZGR 2.0 genes in this pathway |
| SHEDDEN LUNG CANCER GOOD SURVIVAL A12 | 317 | 177 | All SZGR 2.0 genes in this pathway |
| WHITFIELD CELL CYCLE G2 M | 216 | 124 | All SZGR 2.0 genes in this pathway |
| ZWANG CLASS 1 TRANSIENTLY INDUCED BY EGF | 516 | 308 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-101 | 1315 | 1321 | 1A | hsa-miR-101 | UACAGUACUGUGAUAACUGAAG |
| miR-122 | 145 | 151 | 1A | hsa-miR-122a | UGGAGUGUGACAAUGGUGUUUGU |
| miR-125/351 | 470 | 476 | m8 | hsa-miR-125bbrain | UCCCUGAGACCCUAACUUGUGA |
| hsa-miR-125abrain | UCCCUGAGACCCUUUAACCUGUG | ||||
| miR-142-5p | 1360 | 1366 | m8 | hsa-miR-142-5p | CAUAAAGUAGAAAGCACUAC |
| miR-144 | 1315 | 1321 | 1A | hsa-miR-144 | UACAGUAUAGAUGAUGUACUAG |
| miR-150 | 2036 | 2042 | m8 | hsa-miR-150 | UCUCCCAACCCUUGUACCAGUG |
| miR-186 | 69 | 75 | 1A | hsa-miR-186 | CAAAGAAUUCUCCUUUUGGGCUU |
| miR-200bc/429 | 1384 | 1390 | m8 | hsa-miR-200b | UAAUACUGCCUGGUAAUGAUGAC |
| hsa-miR-200c | UAAUACUGCCGGGUAAUGAUGG | ||||
| hsa-miR-429 | UAAUACUGUCUGGUAAAACCGU | ||||
| hsa-miR-200b | UAAUACUGCCUGGUAAUGAUGAC | ||||
| hsa-miR-200c | UAAUACUGCCGGGUAAUGAUGG | ||||
| hsa-miR-429 | UAAUACUGUCUGGUAAAACCGU | ||||
| miR-22 | 1310 | 1316 | 1A | hsa-miR-22brain | AAGCUGCCAGUUGAAGAACUGU |
| miR-28 | 531 | 537 | m8 | hsa-miR-28brain | AAGGAGCUCACAGUCUAUUGAG |
| miR-377 | 2021 | 2027 | m8 | hsa-miR-377 | AUCACACAAAGGCAACUUUUGU |
| miR-485-5p | 398 | 404 | m8 | hsa-miR-485-5p | AGAGGCUGGCCGUGAUGAAUUC |
| miR-486 | 1401 | 1407 | m8 | hsa-miR-486 | UCCUGUACUGAGCUGCCCCGAG |
| miR-7 | 659 | 665 | m8 | hsa-miR-7SZ | UGGAAGACUAGUGAUUUUGUUG |
| miR-9 | 153 | 159 | 1A | hsa-miR-9SZ | UCUUUGGUUAUCUAGCUGUAUGA |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.