Gene Page: TAP2
Summary ?
| GeneID | 6891 |
| Symbol | TAP2 |
| Synonyms | ABC18|ABCB3|APT2|D6S217E|PSF-2|PSF2|RING11 |
| Description | transporter 2, ATP-binding cassette, sub-family B (MDR/TAP) |
| Reference | MIM:170261|HGNC:HGNC:44|Ensembl:ENSG00000204267|HPRD:01360|Vega:OTTHUMG00000031068 |
| Gene type | protein-coding |
| Map location | 6p21.3 |
| Pascal p-value | 6.547E-7 |
| Fetal beta | -0.693 |
| eGene | Caudate basal ganglia Cerebellar Hemisphere Cerebellum Cortex Frontal Cortex BA9 Hippocampus Nucleus accumbens basal ganglia Putamen basal ganglia Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.033 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs2621366 | 6 | 32768980 | TAP2 | ENSG00000204267.9 | 7.641E-7 | 0 | 27831 | gtex_brain_putamen_basal |
| rs2621362 | 6 | 32769536 | TAP2 | ENSG00000204267.9 | 7.641E-7 | 0 | 27275 | gtex_brain_putamen_basal |
| rs2219893 | 6 | 32769663 | TAP2 | ENSG00000204267.9 | 7.641E-7 | 0 | 27148 | gtex_brain_putamen_basal |
| rs2199874 | 6 | 32769926 | TAP2 | ENSG00000204267.9 | 7.641E-7 | 0 | 26885 | gtex_brain_putamen_basal |
| rs199560253 | 6 | 32771452 | TAP2 | ENSG00000204267.9 | 4.706E-7 | 0 | 25359 | gtex_brain_putamen_basal |
| rs375727133 | 6 | 32771847 | TAP2 | ENSG00000204267.9 | 1.241E-6 | 0 | 24964 | gtex_brain_putamen_basal |
| rs2071473 | 6 | 32782605 | TAP2 | ENSG00000204267.9 | 1.077E-7 | 0 | 14206 | gtex_brain_putamen_basal |
| rs2857106 | 6 | 32787570 | TAP2 | ENSG00000204267.9 | 1.18E-7 | 0 | 9241 | gtex_brain_putamen_basal |
| rs2621323 | 6 | 32788707 | TAP2 | ENSG00000204267.9 | 7.433E-8 | 0 | 8104 | gtex_brain_putamen_basal |
| rs2621322 | 6 | 32788712 | TAP2 | ENSG00000204267.9 | 1.18E-7 | 0 | 8099 | gtex_brain_putamen_basal |
| rs2621321 | 6 | 32789480 | TAP2 | ENSG00000204267.9 | 5.991E-7 | 0 | 7331 | gtex_brain_putamen_basal |
| rs2857104 | 6 | 32790167 | TAP2 | ENSG00000204267.9 | 5.991E-7 | 0 | 6644 | gtex_brain_putamen_basal |
| rs2857103 | 6 | 32791299 | TAP2 | ENSG00000204267.9 | 3.09E-8 | 0 | 5512 | gtex_brain_putamen_basal |
| rs2856993 | 6 | 32791403 | TAP2 | ENSG00000204267.9 | 1.18E-7 | 0 | 5408 | gtex_brain_putamen_basal |
| rs13501 | 6 | 32793523 | TAP2 | ENSG00000204267.9 | 3.09E-8 | 0 | 3288 | gtex_brain_putamen_basal |
| rs2857101 | 6 | 32794676 | TAP2 | ENSG00000204267.9 | 5.991E-7 | 0 | 2135 | gtex_brain_putamen_basal |
| rs241456 | 6 | 32795965 | TAP2 | ENSG00000204267.9 | 1.55E-7 | 0 | 846 | gtex_brain_putamen_basal |
| rs241455 | 6 | 32796019 | TAP2 | ENSG00000204267.9 | 1.55E-7 | 0 | 792 | gtex_brain_putamen_basal |
| rs2071546 | 6 | 32796057 | TAP2 | ENSG00000204267.9 | 1.55E-7 | 0 | 754 | gtex_brain_putamen_basal |
| rs2071545 | 6 | 32796058 | TAP2 | ENSG00000204267.9 | 1.5E-7 | 0 | 753 | gtex_brain_putamen_basal |
| rs241454 | 6 | 32796144 | TAP2 | ENSG00000204267.9 | 1.55E-7 | 0 | 667 | gtex_brain_putamen_basal |
| rs241453 | 6 | 32796226 | TAP2 | ENSG00000204267.9 | 3.845E-7 | 0 | 585 | gtex_brain_putamen_basal |
| rs241452 | 6 | 32796346 | TAP2 | ENSG00000204267.9 | 1.55E-7 | 0 | 465 | gtex_brain_putamen_basal |
| rs114247673 | 6 | 32796466 | TAP2 | ENSG00000204267.9 | 2.053E-7 | 0 | 345 | gtex_brain_putamen_basal |
| rs241451 | 6 | 32796480 | TAP2 | ENSG00000204267.9 | 1.988E-8 | 0 | 331 | gtex_brain_putamen_basal |
| rs17034 | 6 | 32796521 | TAP2 | ENSG00000204267.9 | 2.933E-7 | 0 | 290 | gtex_brain_putamen_basal |
| rs241449 | 6 | 32796653 | TAP2 | ENSG00000204267.9 | 1.551E-7 | 0 | 158 | gtex_brain_putamen_basal |
| rs241448 | 6 | 32796685 | TAP2 | ENSG00000204267.9 | 1.551E-7 | 0 | 126 | gtex_brain_putamen_basal |
| rs241447 | 6 | 32796751 | TAP2 | ENSG00000204267.9 | 1.552E-7 | 0 | 60 | gtex_brain_putamen_basal |
| rs241446 | 6 | 32796967 | TAP2 | ENSG00000204267.9 | 3.845E-7 | 0 | -156 | gtex_brain_putamen_basal |
| rs241445 | 6 | 32797072 | TAP2 | ENSG00000204267.9 | 3.845E-7 | 0 | -261 | gtex_brain_putamen_basal |
| rs241444 | 6 | 32797109 | TAP2 | ENSG00000204267.9 | 3.845E-7 | 0 | -298 | gtex_brain_putamen_basal |
| rs241443 | 6 | 32797115 | TAP2 | ENSG00000204267.9 | 6.193E-8 | 0 | -304 | gtex_brain_putamen_basal |
| rs241442 | 6 | 32797168 | TAP2 | ENSG00000204267.9 | 3.845E-7 | 0 | -357 | gtex_brain_putamen_basal |
| rs241441 | 6 | 32797297 | TAP2 | ENSG00000204267.9 | 4.055E-7 | 0 | -486 | gtex_brain_putamen_basal |
| rs241440 | 6 | 32797361 | TAP2 | ENSG00000204267.9 | 3.845E-7 | 0 | -550 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
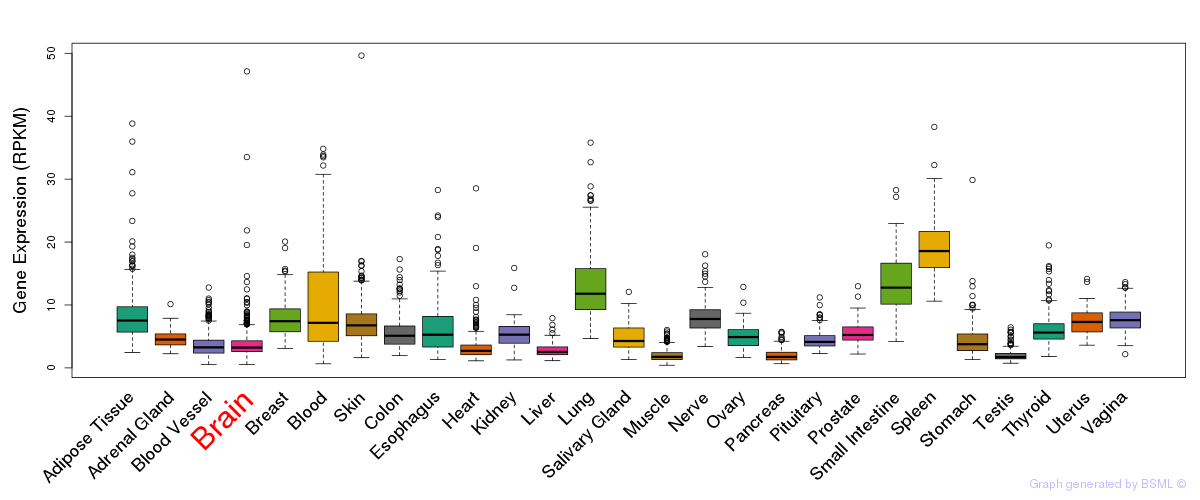
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0005524 | ATP binding | NAS | 12047747 | |
| GO:0004409 | homoaconitate hydratase activity | NAS | 1946428 | |
| GO:0005215 | transporter activity | IDA | 10835348 | |
| GO:0005215 | transporter activity | IEA | - | |
| GO:0015433 | peptide antigen-transporting ATPase activity | NAS | 12047747 | |
| GO:0016887 | ATPase activity | IEA | - | |
| GO:0042288 | MHC class I protein binding | NAS | 12047747 | |
| GO:0042301 | phosphate binding | NAS | 11532960 | |
| GO:0015198 | oligopeptide transporter activity | IEA | - | |
| GO:0015197 | peptide transporter activity | IEA | - | |
| GO:0042605 | peptide antigen binding | NAS | 11133832 | |
| GO:0042626 | ATPase activity, coupled to transmembrane movement of substances | IEA | - | |
| GO:0046982 | protein heterodimerization activity | IPI | 11133832 | |
| GO:0046982 | protein heterodimerization activity | ISS | - | |
| GO:0046980 | tapasin binding | IPI | 12047747 | |
| GO:0046980 | tapasin binding | ISS | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006461 | protein complex assembly | NAS | 12047747 | |
| GO:0019885 | antigen processing and presentation of endogenous peptide antigen via MHC class I | NAS | 11532960 | |
| GO:0006955 | immune response | IEA | - | |
| GO:0006857 | oligopeptide transport | IEA | - | |
| GO:0006886 | intracellular protein transport | IMP | 11133832 | |
| GO:0006886 | intracellular protein transport | ISS | - | |
| GO:0015833 | peptide transport | IEA | - | |
| GO:0046967 | cytosol to ER transport | NAS | 11133832 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005829 | cytosol | NAS | 11133832 | |
| GO:0005788 | endoplasmic reticulum lumen | IMP | 11133832 | |
| GO:0005788 | endoplasmic reticulum lumen | ISS | - | |
| GO:0005789 | endoplasmic reticulum membrane | IEA | - | |
| GO:0005634 | nucleus | IDA | 18029348 | |
| GO:0005783 | endoplasmic reticulum | IDA | 18029348 | |
| GO:0005783 | endoplasmic reticulum | IEA | - | |
| GO:0016020 | membrane | IEA | - | |
| GO:0016021 | integral to membrane | IEA | - | |
| GO:0016021 | integral to membrane | NAS | 1946428 | |
| GO:0042825 | TAP complex | NAS | 12202157 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG ABC TRANSPORTERS | 44 | 29 | All SZGR 2.0 genes in this pathway |
| KEGG ANTIGEN PROCESSING AND PRESENTATION | 89 | 65 | All SZGR 2.0 genes in this pathway |
| KEGG PRIMARY IMMUNODEFICIENCY | 35 | 28 | All SZGR 2.0 genes in this pathway |
| REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | 76 | 48 | All SZGR 2.0 genes in this pathway |
| REACTOME ER PHAGOSOME PATHWAY | 61 | 40 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME ADAPTIVE IMMUNE SYSTEM | 539 | 350 | All SZGR 2.0 genes in this pathway |
| REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | 21 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME CLASS I MHC MEDIATED ANTIGEN PROCESSING PRESENTATION | 251 | 156 | All SZGR 2.0 genes in this pathway |
| HOLLMANN APOPTOSIS VIA CD40 DN | 267 | 178 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS BASAL DN | 455 | 304 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS MESENCHYMAL DN | 460 | 312 | All SZGR 2.0 genes in this pathway |
| BORCZUK MALIGNANT MESOTHELIOMA DN | 104 | 59 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC UP | 722 | 443 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY SERUM DEPRIVATION UP | 552 | 347 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND SERUM DEPRIVATION UP | 211 | 136 | All SZGR 2.0 genes in this pathway |
| RICKMAN METASTASIS DN | 261 | 155 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL FLI1 | 9 | 8 | All SZGR 2.0 genes in this pathway |
| ZHAN MULTIPLE MYELOMA CD1 AND CD2 DN | 51 | 32 | All SZGR 2.0 genes in this pathway |
| JAIN NFKB SIGNALING | 75 | 44 | All SZGR 2.0 genes in this pathway |
| RUAN RESPONSE TO TNF UP | 12 | 10 | All SZGR 2.0 genes in this pathway |
| WANG SMARCE1 TARGETS DN | 371 | 218 | All SZGR 2.0 genes in this pathway |
| RUAN RESPONSE TO TNF TROGLITAZONE UP | 17 | 11 | All SZGR 2.0 genes in this pathway |
| SATO SILENCED BY METHYLATION IN PANCREATIC CANCER 1 | 419 | 273 | All SZGR 2.0 genes in this pathway |
| PELLICCIOTTA HDAC IN ANTIGEN PRESENTATION DN | 49 | 38 | All SZGR 2.0 genes in this pathway |
| WALLACE PROSTATE CANCER RACE UP | 299 | 167 | All SZGR 2.0 genes in this pathway |
| RIZKI TUMOR INVASIVENESS 3D UP | 210 | 124 | All SZGR 2.0 genes in this pathway |
| ICHIBA GRAFT VERSUS HOST DISEASE D7 UP | 107 | 67 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| WUNDER INFLAMMATORY RESPONSE AND CHOLESTEROL UP | 58 | 38 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA CHEMOTAXIS DN | 457 | 302 | All SZGR 2.0 genes in this pathway |
| LU EZH2 TARGETS DN | 414 | 237 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP B | 549 | 316 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS UP | 745 | 475 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS UP | 682 | 433 | All SZGR 2.0 genes in this pathway |
| HUANG GATA2 TARGETS UP | 149 | 96 | All SZGR 2.0 genes in this pathway |
| HOLLEMAN ASPARAGINASE RESISTANCE ALL UP | 22 | 14 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-370 | 1676 | 1682 | m8 | hsa-miR-370brain | GCCUGCUGGGGUGGAACCUGG |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.