Gene Page: TF
Summary ?
| GeneID | 7018 |
| Symbol | TF |
| Synonyms | HEL-S-71p|PRO1557|PRO2086|TFQTL1 |
| Description | transferrin |
| Reference | MIM:190000|HGNC:HGNC:11740|Ensembl:ENSG00000091513|HPRD:01811|Vega:OTTHUMG00000150356 |
| Gene type | protein-coding |
| Map location | 3q22.1 |
| Pascal p-value | 0.351 |
| Sherlock p-value | 0.241 |
| Fetal beta | -3.71 |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| CV:PheWAS | Phenome-wide association studies (PheWAS) | 157 SNPs associated with schizophrenia | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Association | A combined odds ratio method (Sun et al. 2008), association studies | 1 | Link to SZGene |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenics,schizophrenias | Click to show details |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0982 |
Section I. Genetics and epigenetics annotation
 CV:PheWAS
CV:PheWAS
| SNP ID | Chromosome | Position | Allele | P | Function | Gene | Up/Down Distance |
|---|---|---|---|---|---|---|---|
| rs1867504 | 3 | 133410661 | null | 1.679 | TF | null |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs7830259 | chr8 | 14087643 | TF | 7018 | 0.19 | trans |
Section II. Transcriptome annotation
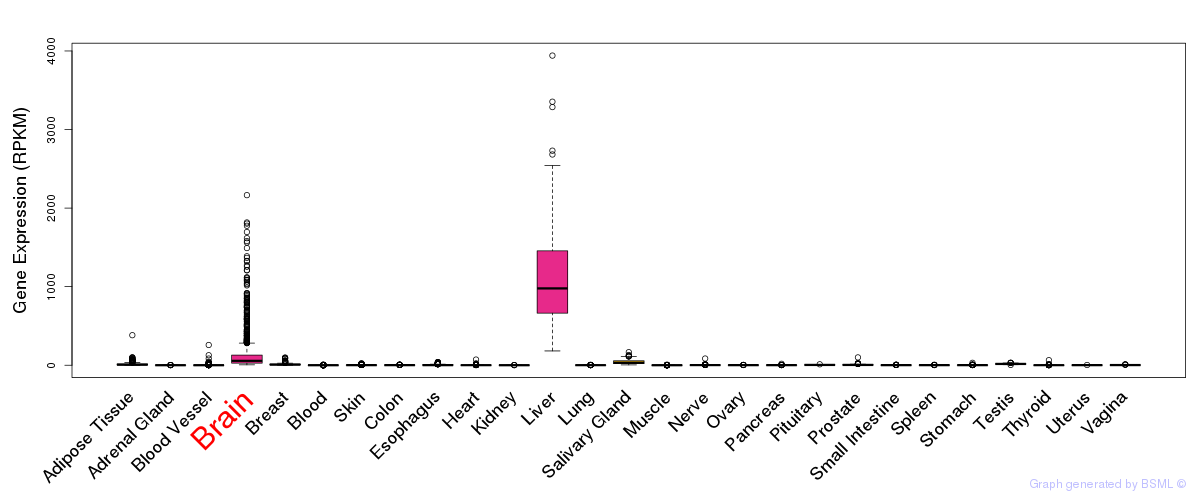
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| HADHB | 0.84 | 0.85 |
| CYP2J2 | 0.84 | 0.85 |
| HSPB8 | 0.82 | 0.86 |
| CSRP1 | 0.81 | 0.83 |
| SDC4 | 0.81 | 0.82 |
| TMBIM1 | 0.80 | 0.86 |
| CKMT2 | 0.80 | 0.75 |
| EFEMP1 | 0.80 | 0.78 |
| STOM | 0.79 | 0.82 |
| GRAMD3 | 0.79 | 0.84 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| NKIRAS2 | -0.58 | -0.55 |
| CRMP1 | -0.57 | -0.57 |
| GPRIN1 | -0.57 | -0.58 |
| ZNF821 | -0.56 | -0.56 |
| C19orf57 | -0.56 | -0.57 |
| NR2C2AP | -0.56 | -0.62 |
| MPP3 | -0.56 | -0.58 |
| AC011491.1 | -0.56 | -0.65 |
| GMIP | -0.56 | -0.57 |
| HN1 | -0.55 | -0.59 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0008199 | ferric iron binding | IEA | - | |
| GO:0046872 | metal ion binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006826 | iron ion transport | IEA | - | |
| GO:0006811 | ion transport | IEA | - | |
| GO:0006879 | cellular iron ion homeostasis | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005576 | extracellular region | IEA | - | |
| GO:0005576 | extracellular region | NAS | 14718574 | |
| GO:0005739 | mitochondrion | IDA | 18029348 | |
| GO:0005768 | endosome | IEA | - | |
| GO:0030139 | endocytic vesicle | IDA | 15229288 |15292400 | |
| GO:0030139 | endocytic vesicle | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| CALR | CRT | FLJ26680 | RO | SSA | cC1qR | calreticulin | - | HPRD,BioGRID | 9312001 |
| CANX | CNX | FLJ26570 | IP90 | P90 | calnexin | - | HPRD,BioGRID | 9312001 |
| CUBN | FLJ90055 | FLJ90747 | IFCR | MGA1 | gp280 | cubilin (intrinsic factor-cobalamin receptor) | Transferrin interacts with Cubilin. | BIND | 11606717 |
| FNBP1 | FBP17 | KIAA0554 | MGC126804 | formin binding protein 1 | Co-localization | BioGRID | 15252009 |
| IGF1 | IGFI | insulin-like growth factor 1 (somatomedin C) | Reconstituted Complex | BioGRID | 11749962 |
| IGF2 | C11orf43 | FLJ22066 | FLJ44734 | INSIGF | pp9974 | insulin-like growth factor 2 (somatomedin A) | Reconstituted Complex | BioGRID | 11749962 |
| IGFBP1 | AFBP | IBP1 | IGF-BP25 | PP12 | hIGFBP-1 | insulin-like growth factor binding protein 1 | - | HPRD | 11297622 |
| IGFBP2 | IBP2 | IGF-BP53 | insulin-like growth factor binding protein 2, 36kDa | - | HPRD | 11297622 |
| IGFBP3 | BP-53 | IBP3 | insulin-like growth factor binding protein 3 | - | HPRD,BioGRID | 11297622 |11749962 |
| IGFBP4 | BP-4 | HT29-IGFBP | IBP4 | IGFBP-4 | insulin-like growth factor binding protein 4 | - | HPRD | 11297622 |
| IGFBP5 | IBP5 | insulin-like growth factor binding protein 5 | - | HPRD | 11297622 |
| IGFBP6 | IBP6 | insulin-like growth factor binding protein 6 | - | HPRD | 11297622 |
| TFR2 | HFE3 | MGC126368 | TFRC2 | transferrin receptor 2 | - | HPRD,BioGRID | 11027676 |
| TFRC | CD71 | TFR | TFR1 | TRFR | transferrin receptor (p90, CD71) | Tf interacts with TfR. | BIND | 15525538 |
| TFRC | CD71 | TFR | TFR1 | TRFR | transferrin receptor (p90, CD71) | Reconstituted Complex | BioGRID | 11027676 |
| TFRC | CD71 | TFR | TFR1 | TRFR | transferrin receptor (p90, CD71) | - | HPRD | 6268632 |
| TUBB3 | MC1R | TUBB4 | beta-4 | tubulin, beta 3 | Two-hybrid | BioGRID | 16169070 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| ST WNT CA2 CYCLIC GMP PATHWAY | 20 | 14 | All SZGR 2.0 genes in this pathway |
| PID EPHB FWD PATHWAY | 40 | 38 | All SZGR 2.0 genes in this pathway |
| PID HIF1 TFPATHWAY | 66 | 52 | All SZGR 2.0 genes in this pathway |
| REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | 89 | 56 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSMEMBRANE TRANSPORT OF SMALL MOLECULES | 413 | 270 | All SZGR 2.0 genes in this pathway |
| REACTOME IRON UPTAKE AND TRANSPORT | 36 | 18 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | 25 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME HEMOSTASIS | 466 | 331 | All SZGR 2.0 genes in this pathway |
| REACTOME PLATELET ACTIVATION SIGNALING AND AGGREGATION | 208 | 138 | All SZGR 2.0 genes in this pathway |
| PARENT MTOR SIGNALING UP | 567 | 375 | All SZGR 2.0 genes in this pathway |
| SENGUPTA NASOPHARYNGEAL CARCINOMA WITH LMP1 DN | 175 | 82 | All SZGR 2.0 genes in this pathway |
| TURASHVILI BREAST DUCTAL CARCINOMA VS DUCTAL NORMAL DN | 198 | 110 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| HATADA METHYLATED IN LUNG CANCER UP | 390 | 236 | All SZGR 2.0 genes in this pathway |
| MANTOVANI VIRAL GPCR SIGNALING DN | 49 | 37 | All SZGR 2.0 genes in this pathway |
| BERTUCCI INVASIVE CARCINOMA DUCTAL VS LOBULAR DN | 46 | 34 | All SZGR 2.0 genes in this pathway |
| RICKMAN HEAD AND NECK CANCER D | 37 | 14 | All SZGR 2.0 genes in this pathway |
| COLIN PILOCYTIC ASTROCYTOMA VS GLIOBLASTOMA UP | 35 | 32 | All SZGR 2.0 genes in this pathway |
| SHEN SMARCA2 TARGETS DN | 357 | 212 | All SZGR 2.0 genes in this pathway |
| CERVERA SDHB TARGETS 2 | 114 | 76 | All SZGR 2.0 genes in this pathway |
| ASTON MAJOR DEPRESSIVE DISORDER DN | 160 | 110 | All SZGR 2.0 genes in this pathway |
| MATSUDA NATURAL KILLER DIFFERENTIATION | 475 | 313 | All SZGR 2.0 genes in this pathway |
| HSIAO LIVER SPECIFIC GENES | 244 | 154 | All SZGR 2.0 genes in this pathway |
| BROWN MYELOID CELL DEVELOPMENT DN | 129 | 86 | All SZGR 2.0 genes in this pathway |
| HESS TARGETS OF HOXA9 AND MEIS1 DN | 77 | 48 | All SZGR 2.0 genes in this pathway |
| HARRIS HYPOXIA | 81 | 64 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| SEMENZA HIF1 TARGETS | 36 | 32 | All SZGR 2.0 genes in this pathway |
| WANG CISPLATIN RESPONSE AND XPC UP | 202 | 115 | All SZGR 2.0 genes in this pathway |
| CHENG RESPONSE TO NICKEL ACETATE | 45 | 29 | All SZGR 2.0 genes in this pathway |
| ZHONG SECRETOME OF LUNG CANCER AND ENDOTHELIUM | 66 | 47 | All SZGR 2.0 genes in this pathway |
| ZHONG SECRETOME OF LUNG CANCER AND FIBROBLAST | 132 | 93 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS UP | 673 | 430 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS UP | 602 | 364 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS UP | 601 | 369 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER NORMAL LIKE UP | 476 | 285 | All SZGR 2.0 genes in this pathway |
| CADWELL ATG16L1 TARGETS UP | 93 | 56 | All SZGR 2.0 genes in this pathway |
| BREDEMEYER RAG SIGNALING NOT VIA ATM DN | 57 | 34 | All SZGR 2.0 genes in this pathway |
| WINTER HYPOXIA METAGENE | 242 | 168 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| POOLA INVASIVE BREAST CANCER DN | 134 | 83 | All SZGR 2.0 genes in this pathway |
| BOYAULT LIVER CANCER SUBCLASS G3 DN | 51 | 32 | All SZGR 2.0 genes in this pathway |
| LI INDUCED T TO NATURAL KILLER UP | 307 | 182 | All SZGR 2.0 genes in this pathway |
| CHYLA CBFA2T3 TARGETS UP | 387 | 225 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS UP | 504 | 321 | All SZGR 2.0 genes in this pathway |
| VANDESLUIS COMMD1 TARGETS GROUP 3 UP | 89 | 50 | All SZGR 2.0 genes in this pathway |
| YANG BCL3 TARGETS UP | 364 | 236 | All SZGR 2.0 genes in this pathway |