Gene Page: TRAF2
Summary ?
| GeneID | 7186 |
| Symbol | TRAF2 |
| Synonyms | MGC:45012|TRAP|TRAP3 |
| Description | TNF receptor associated factor 2 |
| Reference | MIM:601895|HGNC:HGNC:12032|Ensembl:ENSG00000127191|HPRD:03538|Vega:OTTHUMG00000020952 |
| Gene type | protein-coding |
| Map location | 9q34 |
| Pascal p-value | 0.583 |
| Sherlock p-value | 0.674 |
| Support | Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.2666 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
General gene expression (GTEx)
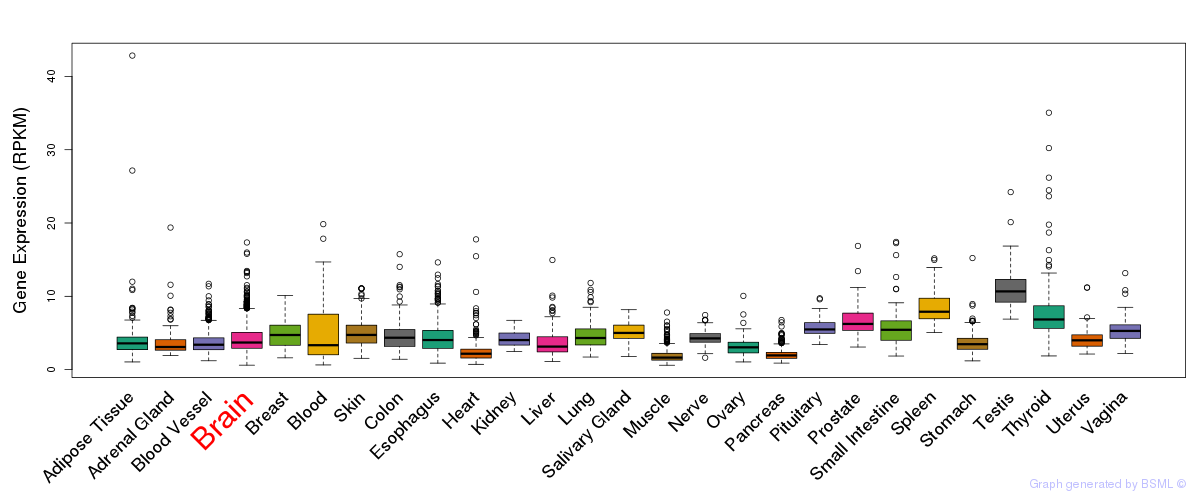
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004871 | signal transducer activity | NAS | 8702708 | |
| GO:0005515 | protein binding | IPI | 11466612 |11777919 | |
| GO:0008270 | zinc ion binding | IEA | - | |
| GO:0046872 | metal ion binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0002726 | positive regulation of T cell cytokine production | IMP | 15125833 | |
| GO:0006461 | protein complex assembly | TAS | 8702708 | |
| GO:0007250 | activation of NF-kappaB-inducing kinase activity | IMP | 15125833 | |
| GO:0007165 | signal transduction | TAS | 8702708 | |
| GO:0008633 | activation of pro-apoptotic gene products | EXP | 14644197 | |
| GO:0042981 | regulation of apoptosis | IEA | - | |
| GO:0050870 | positive regulation of T cell activation | IC | 15125833 | |
| GO:0032743 | positive regulation of interleukin-2 production | IMP | 15125833 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005829 | cytosol | EXP | 7758105 |12887920 | |
| GO:0005737 | cytoplasm | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| AASDHPPT | AASD-PPT | CGI-80 | DKFZp566E2346 | LYS2 | LYS5 | aminoadipate-semialdehyde dehydrogenase-phosphopantetheinyl transferase | Two-hybrid | BioGRID | 16189514 |
| ABLIM1 | ABLIM | DKFZp781D0148 | FLJ14564 | KIAA0059 | LIMAB1 | LIMATIN | MGC1224 | actin binding LIM protein 1 | Two-hybrid | BioGRID | 16189514 |
| AFF4 | AF5Q31 | MCEF | MGC75036 | AF4/FMR2 family, member 4 | Two-hybrid | BioGRID | 16189514 |
| ANKRD26L1 | MGC12538 | ankyrin repeat domain 26-like 1 | Two-hybrid | BioGRID | 16189514 |
| APEX2 | APE2 | APEXL2 | XTH2 | APEX nuclease (apurinic/apyrimidinic endonuclease) 2 | Two-hybrid | BioGRID | 16189514 |
| APIP | APIP2 | CGI-29 | MMRP19 | dJ179L10.2 | APAF1 interacting protein | - | HPRD | 14743216 |
| APPL1 | APPL | DIP13alpha | adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 1 | Two-hybrid | BioGRID | 16189514 |
| BAHD1 | KIAA0945 | bromo adjacent homology domain containing 1 | Two-hybrid | BioGRID | 16189514 |
| BANP | BEND1 | DKFZp761H172 | FLJ10177 | FLJ20538 | SMAR1 | SMARBP1 | BTG3 associated nuclear protein | Two-hybrid | BioGRID | 16189514 |
| BCL10 | CARMEN | CIPER | CLAP | c-E10 | mE10 | B-cell CLL/lymphoma 10 | - | HPRD,BioGRID | 10753917 |
| BIRC2 | API1 | HIAP2 | Hiap-2 | MIHB | RNF48 | cIAP1 | baculoviral IAP repeat-containing 2 | c-IAP1 interacts with and ubiquitinates TRAF2. | BIND | 11907583 |
| BIRC2 | API1 | HIAP2 | Hiap-2 | MIHB | RNF48 | cIAP1 | baculoviral IAP repeat-containing 2 | - | HPRD | 8643514 |9384571 |11907583 |
| BIRC2 | API1 | HIAP2 | Hiap-2 | MIHB | RNF48 | cIAP1 | baculoviral IAP repeat-containing 2 | Affinity Capture-Western in vitro Reconstituted Complex Two-hybrid | BioGRID | 8643514 |8943045 |9384571 |11907583 |16189514 |
| BIRC3 | AIP1 | API2 | CIAP2 | HAIP1 | HIAP1 | MALT2 | MIHC | RNF49 | baculoviral IAP repeat-containing 3 | - | HPRD | 8643514 |11907583 |
| BIRC3 | AIP1 | API2 | CIAP2 | HAIP1 | HIAP1 | MALT2 | MIHC | RNF49 | baculoviral IAP repeat-containing 3 | in vitro Reconstituted Complex Two-hybrid | BioGRID | 8643514 |9384571 |11907583 |
| BIRC3 | AIP1 | API2 | CIAP2 | HAIP1 | HIAP1 | MALT2 | MIHC | RNF49 | baculoviral IAP repeat-containing 3 | c-IAP2 interacts with TRAF2. | BIND | 11907583 |
| C16orf48 | DAKV6410 | DKFZp434A1319 | chromosome 16 open reading frame 48 | Two-hybrid | BioGRID | 16189514 |
| CALCOCO2 | MGC17318 | NDP52 | calcium binding and coiled-coil domain 2 | Two-hybrid | BioGRID | 16189514 |
| CASP12 | CASP12P1 | caspase 12 (gene/pseudogene) | Affinity Capture-Western | BioGRID | 11278723 |
| CASP8 | ALPS2B | CAP4 | FLICE | FLJ17672 | MACH | MCH5 | MGC78473 | caspase 8, apoptosis-related cysteine peptidase | Affinity Capture-Western | BioGRID | 12887920 |
| CASP8AP2 | CED-4 | FLASH | FLJ11208 | KIAA1315 | RIP25 | caspase 8 associated protein 2 | - | HPRD,BioGRID | 11340079 |
| CAV1 | CAV | MSTP085 | VIP21 | caveolin 1, caveolae protein, 22kDa | - | HPRD,BioGRID | 11112773 |11805080 |
| CCDC130 | MGC10471 | coiled-coil domain containing 130 | Two-hybrid | BioGRID | 16189514 |
| CCDC33 | FLJ23168 | FLJ32855 | MGC34145 | coiled-coil domain containing 33 | Two-hybrid | BioGRID | 16189514 |
| CD27 | MGC20393 | S152 | T14 | TNFRSF7 | Tp55 | CD27 molecule | - | HPRD,BioGRID | 9582383 |
| CD40 | Bp50 | CDW40 | MGC9013 | TNFRSF5 | p50 | CD40 molecule, TNF receptor superfamily member 5 | - | HPRD,BioGRID | 10411888 |
| CD40 | Bp50 | CDW40 | MGC9013 | TNFRSF5 | p50 | CD40 molecule, TNF receptor superfamily member 5 | CD40 interacts with TRAF2. | BIND | 10352240 |
| CDCA3 | GRCC8 | MGC2577 | TOME-1 | cell division cycle associated 3 | Two-hybrid | BioGRID | 16189514 |
| CDKN1B | CDKN4 | KIP1 | MEN1B | MEN4 | P27KIP1 | cyclin-dependent kinase inhibitor 1B (p27, Kip1) | Two-hybrid | BioGRID | 16189514 |
| CFLAR | CASH | CASP8AP1 | CLARP | Casper | FLAME | FLAME-1 | FLAME1 | FLIP | I-FLICE | MRIT | c-FLIP | c-FLIPL | c-FLIPR | c-FLIPS | CASP8 and FADD-like apoptosis regulator | - | HPRD,BioGRID | 9208847 |
| CHMP2B | CHMP2.5 | DKFZp564O123 | DMT1 | VPS2-2 | VPS2B | chromatin modifying protein 2B | Two-hybrid | BioGRID | 16189514 |
| CHUK | IKBKA | IKK-alpha | IKK1 | IKKA | NFKBIKA | TCF16 | conserved helix-loop-helix ubiquitous kinase | - | HPRD,BioGRID | 11359906 |
| CRY1 | PHLL1 | cryptochrome 1 (photolyase-like) | Two-hybrid | BioGRID | 16189514 |
| CYLD | CDMT | CYLD1 | CYLDI | EAC | FLJ20180 | FLJ31664 | FLJ78684 | HSPC057 | KIAA0849 | MFT | MFT1 | SBS | TEM | USPL2 | cylindromatosis (turban tumor syndrome) | CYLD interacts with TRAF2. | BIND | 12917691 |
| DLGAP5 | DLG1 | DLG7 | HURP | KIAA0008 | discs, large (Drosophila) homolog-associated protein 5 | Two-hybrid | BioGRID | 16189514 |
| DTNB | MGC17163 | MGC57126 | dystrobrevin, beta | Two-hybrid | BioGRID | 16189514 |
| DVL2 | - | dishevelled, dsh homolog 2 (Drosophila) | Two-hybrid | BioGRID | 16189514 |
| EDARADD | ED3 | EDA3 | EDAR-associated death domain | Affinity Capture-Western | BioGRID | 11882293 |
| ERN1 | FLJ30999 | IRE1 | IRE1P | MGC163277 | MGC163279 | endoplasmic reticulum to nucleus signaling 1 | - | HPRD,BioGRID | 10650002 |12843613 |
| FAM107A | DRR1 | FLJ30158 | FLJ45473 | TU3A | family with sequence similarity 107, member A | Two-hybrid | BioGRID | 16189514 |
| FAM120B | CCPG | FLJ55614 | KIAA0183 | KIAA1838 | PGCC1 | dJ894D12.1 | family with sequence similarity 120B | Two-hybrid | BioGRID | 16189514 |
| FATE1 | FATE | fetal and adult testis expressed 1 | Two-hybrid | BioGRID | 16189514 |
| FBF1 | Alb | FBF-1 | FLJ00103 | Fas (TNFRSF6) binding factor 1 | Two-hybrid | BioGRID | 16189514 |
| FBXO28 | FLJ10766 | Fbx28 | KIAA0483 | F-box protein 28 | Two-hybrid | BioGRID | 16189514 |
| FLNA | ABP-280 | ABPX | DKFZp434P031 | FLN | FLN1 | FMD | MNS | NHBP | OPD | OPD1 | OPD2 | filamin A, alpha (actin binding protein 280) | - | HPRD,BioGRID | 10617615 |
| FXR2 | FMR1L2 | fragile X mental retardation, autosomal homolog 2 | Two-hybrid | BioGRID | 16189514 |
| GORASP2 | DKFZp434D156 | FLJ13139 | GOLPH6 | GRASP55 | GRS2 | p59 | golgi reassembly stacking protein 2, 55kDa | Two-hybrid | BioGRID | 16189514 |
| HIVEP3 | FLJ16752 | KBP-1 | KBP1 | KIAA1555 | KRC | SHN3 | Schnurri-3 | ZAS3 | ZNF40C | human immunodeficiency virus type I enhancer binding protein 3 | - | HPRD,BioGRID | 11804591 |
| HSPA4 | APG-2 | HS24/P52 | MGC131852 | RY | hsp70 | hsp70RY | heat shock 70kDa protein 4 | - | HPRD | 14743216 |
| HSPA8 | HSC54 | HSC70 | HSC71 | HSP71 | HSP73 | HSPA10 | LAP1 | MGC131511 | MGC29929 | NIP71 | heat shock 70kDa protein 8 | - | HPRD | 14743216 |
| IKBKB | FLJ40509 | IKK-beta | IKK2 | IKKB | MGC131801 | NFKBIKB | inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase beta | - | HPRD,BioGRID | 11359906 |
| IL15RA | MGC104179 | interleukin 15 receptor, alpha | - | HPRD,BioGRID | 11064452 |
| IPO11 | RanBP11 | importin 11 | Two-hybrid | BioGRID | 16189514 |
| IRF4 | LSIRF | MUM1 | interferon regulatory factor 4 | Two-hybrid | BioGRID | 16189514 |
| ITPK1 | ITRPK1 | inositol 1,3,4-triphosphate 5/6 kinase | Two-hybrid | BioGRID | 16189514 |
| KIAA0408 | FLJ43995 | RP3-403A15.2 | KIAA0408 | Two-hybrid | BioGRID | 16189514 |
| KIAA1267 | DKFZp686P06109 | DKFZp727C091 | MGC102843 | KIAA1267 | Two-hybrid | BioGRID | 16189514 |
| LNX1 | LNX | MPDZ | PDZRN2 | ligand of numb-protein X 1 | Two-hybrid | BioGRID | 16189514 |
| LOC116349 | - | hypothetical protein BC014011 | Two-hybrid | BioGRID | 16189514 |
| LOC170082 | MGC17403 | TFIIS central domain-containing protein 1 | Two-hybrid | BioGRID | 16189514 |
| LTBR | CD18 | D12S370 | LT-BETA-R | TNF-R-III | TNFCR | TNFR-RP | TNFR2-RP | TNFRSF3 | lymphotoxin beta receptor (TNFR superfamily, member 3) | - | HPRD | 12169272 |
| LTBR | CD18 | D12S370 | LT-BETA-R | TNF-R-III | TNFCR | TNFR-RP | TNFR2-RP | TNFRSF3 | lymphotoxin beta receptor (TNFR superfamily, member 3) | TRAF2 interacts with LTBR. | BIND | 12571250 |
| LTBR | CD18 | D12S370 | LT-BETA-R | TNF-R-III | TNFCR | TNFR-RP | TNFR2-RP | TNFRSF3 | lymphotoxin beta receptor (TNFR superfamily, member 3) | Reconstituted Complex | BioGRID | 9162022 |
| LZTS2 | KIAA1813 | LAPSER1 | leucine zipper, putative tumor suppressor 2 | Two-hybrid | BioGRID | 16189514 |
| MALT1 | DKFZp434L132 | MLT | MLT1 | mucosa associated lymphoid tissue lymphoma translocation gene 1 | TRAF2 interacts with MALT1. | BIND | 15125833 |
| MAP3K1 | MAPKKK1 | MEKK | MEKK1 | mitogen-activated protein kinase kinase kinase 1 | Affinity Capture-Western Co-fractionation | BioGRID | 10346818 |
| MAP3K14 | FTDCR1B | HS | HSNIK | NIK | mitogen-activated protein kinase kinase kinase 14 | - | HPRD,BioGRID | 9020361 |11278268 |
| MAP3K14 | FTDCR1B | HS | HSNIK | NIK | mitogen-activated protein kinase kinase kinase 14 | NIK interacts with TRAF2. This interaction was modeled on a demonstrated interaction between human NIK and TRAF2 from an unspecified species. | BIND | 9275204 |
| MAP3K5 | ASK1 | MAPKKK5 | MEKK5 | mitogen-activated protein kinase kinase kinase 5 | - | HPRD,BioGRID | 9774977 |10523862 |
| MAP3K7 | TAK1 | TGF1a | mitogen-activated protein kinase kinase kinase 7 | Affinity Capture-Western | BioGRID | 14633987 |
| MAP3K7 | TAK1 | TGF1a | mitogen-activated protein kinase kinase kinase 7 | - | HPRD | 14743216 |
| MAP3K7IP1 | 3'-Tab1 | MGC57664 | TAB1 | mitogen-activated protein kinase kinase kinase 7 interacting protein 1 | Affinity Capture-MS | BioGRID | 14743216 |
| MAP3K7IP3 | MGC45404 | NAP1 | TAB3 | mitogen-activated protein kinase kinase kinase 7 interacting protein 3 | - | HPRD,BioGRID | 14633987 |
| MAP3K8 | COT | EST | ESTF | FLJ10486 | TPL2 | Tpl-2 | c-COT | mitogen-activated protein kinase kinase kinase 8 | - | HPRD,BioGRID | 11932422 |
| MAP4K2 | BL44 | GCK | RAB8IP | mitogen-activated protein kinase kinase kinase kinase 2 | - | HPRD,BioGRID | 9712898 |
| MAP4K2 | BL44 | GCK | RAB8IP | mitogen-activated protein kinase kinase kinase kinase 2 | The carboxy-terminal TRAF domain of TRAF2 interacts with the carboxy-terminal domain of GCK. | BIND | 9712898 |
| MAP4K5 | GCKR | KHS | KHS1 | MAPKKKK5 | mitogen-activated protein kinase kinase kinase kinase 5 | - | HPRD,BioGRID | 10477597 |
| MAPK9 | JNK-55 | JNK2 | JNK2A | JNK2ALPHA | JNK2B | JNK2BETA | PRKM9 | SAPK | p54a | p54aSAPK | mitogen-activated protein kinase 9 | - | HPRD | 10490605 |
| MAST2 | FLJ39200 | KIAA0807 | MAST205 | MTSSK | RP4-533D7.1 | microtubule associated serine/threonine kinase 2 | Affinity Capture-Western | BioGRID | 15308666 |
| MIZF | DKFZp434F162 | HiNF-P | ZNF743 | MBD2-interacting zinc finger | Two-hybrid | BioGRID | 16189514 |
| NECAP2 | FLJ10420 | RP4-798A10.1 | NECAP endocytosis associated 2 | Two-hybrid | BioGRID | 16189514 |
| NGFR | CD271 | Gp80-LNGFR | TNFRSF16 | p75(NTR) | p75NTR | nerve growth factor receptor (TNFR superfamily, member 16) | - | HPRD,BioGRID | 10514511 |
| NGFRAP1 | BEX3 | Bex | DXS6984E | HGR74 | NADE | nerve growth factor receptor (TNFRSF16) associated protein 1 | - | HPRD | 10514511 |
| NIF3L1 | ALS2CR1 | CALS-7 | MDS015 | NIF3 NGG1 interacting factor 3-like 1 (S. pombe) | Two-hybrid | BioGRID | 16189514 |
| NRIP1 | FLJ77253 | RIP140 | nuclear receptor interacting protein 1 | Two-hybrid | BioGRID | 16189514 |
| NUDT18 | FLJ22494 | nudix (nucleoside diphosphate linked moiety X)-type motif 18 | Two-hybrid | BioGRID | 16189514 |
| PIN1 | DOD | UBL5 | peptidylprolyl cis/trans isomerase, NIMA-interacting 1 | Two-hybrid | BioGRID | 16189514 |
| PSMF1 | PI31 | proteasome (prosome, macropain) inhibitor subunit 1 (PI31) | Two-hybrid | BioGRID | 16189514 |
| PTPN2 | PTPT | TC-PTP | TCELLPTP | TCPTP | protein tyrosine phosphatase, non-receptor type 2 | PTPN2 (TCPTP) interacts with TRAF2. | BIND | 15696169 |
| QARS | GLNRS | PRO2195 | glutaminyl-tRNA synthetase | Two-hybrid | BioGRID | 16189514 |
| RAD23A | HHR23A | MGC111083 | RAD23 homolog A (S. cerevisiae) | Two-hybrid | BioGRID | 16189514 |
| RB1CC1 | CC1 | DRAGOU14 | FIP200 | RB1-inducible coiled-coil 1 | Affinity Capture-Western | BioGRID | 17015619 |
| RFX3 | MGC87155 | bA32F11.1 | regulatory factor X, 3 (influences HLA class II expression) | Two-hybrid | BioGRID | 16189514 |
| RGS14 | - | regulator of G-protein signaling 14 | Two-hybrid | BioGRID | 16189514 |
| RIBC2 | C22orf11 | RIB43A domain with coiled-coils 2 | Two-hybrid | BioGRID | 16189514 |
| RIPK1 | FLJ39204 | RIP | RIP1 | receptor (TNFRSF)-interacting serine-threonine kinase 1 | - | HPRD,BioGRID | 8612133 |8702708 |
| RIPK1 | FLJ39204 | RIP | RIP1 | receptor (TNFRSF)-interacting serine-threonine kinase 1 | TRAF2 ubiquitinates RIP. | BIND | 15258597 |
| RIPK1 | FLJ39204 | RIP | RIP1 | receptor (TNFRSF)-interacting serine-threonine kinase 1 | TRAF2 interacts with RIP. | BIND | 8702708 |
| RIPK2 | CARD3 | CARDIAK | CCK | GIG30 | RICK | RIP2 | receptor-interacting serine-threonine kinase 2 | - | HPRD,BioGRID | 9705938 |
| RIPK3 | RIP3 | receptor-interacting serine-threonine kinase 3 | - | HPRD,BioGRID | 10339433 |
| RUVBL2 | CGI-46 | ECP51 | INO80J | REPTIN | RVB2 | TIH2 | TIP48 | TIP49B | RuvB-like 2 (E. coli) | - | HPRD | 14743216 |
| SETD5 | DKFZp686J18276 | FLJ10707 | KIAA1757 | SET domain containing 5 | Two-hybrid | BioGRID | 16189514 |
| SIAH2 | hSiah2 | seven in absentia homolog 2 (Drosophila) | - | HPRD,BioGRID | 12411493 |
| SOX30 | - | SRY (sex determining region Y)-box 30 | Two-hybrid | BioGRID | 16189514 |
| SOX9 | CMD1 | CMPD1 | SRA1 | SRY (sex determining region Y)-box 9 | - | HPRD | 12136106 |
| SPATA2 | FLJ13167 | KIAA0757 | PD1 | tamo | spermatogenesis associated 2 | Two-hybrid | BioGRID | 16189514 |
| SPG21 | ACP33 | BM-019 | GL010 | MASPARDIN | MAST | spastic paraplegia 21 (autosomal recessive, Mast syndrome) | Two-hybrid | BioGRID | 16189514 |
| SPHK1 | SPHK | sphingosine kinase 1 | - | HPRD,BioGRID | 11777919 |
| SPRY2 | MGC23039 | hSPRY2 | sprouty homolog 2 (Drosophila) | Two-hybrid | BioGRID | 16189514 |
| TANK | I-TRAF | TRAF2 | TRAF family member-associated NFKB activator | - | HPRD,BioGRID | 8710854 |10581243 |
| TAOK3 | DKFZp666H245 | DPK | FLJ31808 | JIK | MAP3K18 | TAO kinase 3 | - | HPRD,BioGRID | 11278723 |
| TBK1 | FLJ11330 | NAK | T2K | TANK-binding kinase 1 | - | HPRD,BioGRID | 10990461 |
| TCEA2 | TFIIS | transcription elongation factor A (SII), 2 | Two-hybrid | BioGRID | 16189514 |
| TCEB3B | ELOA2 | HsT832 | MGC119351 | TCEB3L | transcription elongation factor B polypeptide 3B (elongin A2) | Two-hybrid | BioGRID | 16189514 |
| THAP7 | MGC10963 | THAP domain containing 7 | Two-hybrid | BioGRID | 16189514 |
| TIFA | MGC20791 | T2BP | T6BP | TIFAA | TRAF-interacting protein with forkhead-associated domain | Two-hybrid | BioGRID | 16189514 |
| TIFA | MGC20791 | T2BP | T6BP | TIFAA | TRAF-interacting protein with forkhead-associated domain | - | HPRD | 11798190 |
| TNF | DIF | TNF-alpha | TNFA | TNFSF2 | tumor necrosis factor (TNF superfamily, member 2) | Affinity Capture-Western | BioGRID | 12887920 |
| TNFAIP3 | A20 | MGC104522 | MGC138687 | MGC138688 | OTUD7C | TNFA1P2 | tumor necrosis factor, alpha-induced protein 3 | - | HPRD,BioGRID | 8692885 |
| TNFRSF11A | CD265 | FEO | LOH18CR1 | ODFR | OFE | OPTB7 | OSTS | PDB2 | RANK | TRANCER | tumor necrosis factor receptor superfamily, member 11a, NFKB activator | Affinity Capture-Western Reconstituted Complex | BioGRID | 9685412 |9852070 |10025951 |10075662 |
| TNFRSF11A | CD265 | FEO | LOH18CR1 | ODFR | OFE | OPTB7 | OSTS | PDB2 | RANK | TRANCER | tumor necrosis factor receptor superfamily, member 11a, NFKB activator | - | HPRD | 9774460 |9852070 |10025951 |
| TNFRSF12A | CD266 | FN14 | TWEAKR | tumor necrosis factor receptor superfamily, member 12A | - | HPRD,BioGRID | 11728344 |
| TNFRSF13B | CD267 | CVID | FLJ39942 | MGC133214 | MGC39952 | TACI | TNFRSF14B | tumor necrosis factor receptor superfamily, member 13B | - | HPRD,BioGRID | 10880535 |
| TNFRSF14 | ATAR | HVEA | HVEM | LIGHTR | TR2 | tumor necrosis factor receptor superfamily, member 14 (herpesvirus entry mediator) | - | HPRD,BioGRID | 9153189 |9162022 |
| TNFRSF17 | BCM | BCMA | CD269 | tumor necrosis factor receptor superfamily, member 17 | - | HPRD,BioGRID | 10903733 |
| TNFRSF18 | AITR | GITR | GITR-D | tumor necrosis factor receptor superfamily, member 18 | - | HPRD,BioGRID | 10037686 |
| TNFRSF19 | TAJ | TAJ-alpha | TRADE | TROY | tumor necrosis factor receptor superfamily, member 19 | - | HPRD,BioGRID | 10809768 |
| TNFRSF1A | CD120a | FPF | MGC19588 | TBP1 | TNF-R | TNF-R-I | TNF-R55 | TNFAR | TNFR1 | TNFR55 | TNFR60 | p55 | p55-R | p60 | tumor necrosis factor receptor superfamily, member 1A | - | HPRD,BioGRID | 8565075 |
| TNFRSF1B | CD120b | TBPII | TNF-R-II | TNF-R75 | TNFBR | TNFR1B | TNFR2 | TNFR80 | p75 | p75TNFR | tumor necrosis factor receptor superfamily, member 1B | TRAF2 interacts with TRAF1. This interaction was modeled on a demonstrated interaction between human TRAF2 and TNF-R2 from an unspecified species. | BIND | 8702708 |
| TNFRSF1B | CD120b | TBPII | TNF-R-II | TNF-R75 | TNFBR | TNFR1B | TNFR2 | TNFR80 | p75 | p75TNFR | tumor necrosis factor receptor superfamily, member 1B | - | HPRD | 7639698 |8702708 |9461607 |11907088 |
| TNFRSF1B | CD120b | TBPII | TNF-R-II | TNF-R75 | TNFBR | TNFR1B | TNFR2 | TNFR80 | p75 | p75TNFR | tumor necrosis factor receptor superfamily, member 1B | Affinity Capture-MS Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 7639698 |8702708 |8710854 |9162022 |11907088 |14743216 |
| TNFRSF4 | ACT35 | CD134 | OX40 | TXGP1L | tumor necrosis factor receptor superfamily, member 4 | - | HPRD,BioGRID | 9418902 |
| TNFRSF8 | CD30 | D1S166E | KI-1 | tumor necrosis factor receptor superfamily, member 8 | - | HPRD | 8943059 |8999898 |
| TNFRSF8 | CD30 | D1S166E | KI-1 | tumor necrosis factor receptor superfamily, member 8 | - | HPRD,BioGRID | 8627180 |8943059 |8999898 |9168896 |9353251 |11971184|8943059 |8999898 |
| TNFRSF8 | CD30 | D1S166E | KI-1 | tumor necrosis factor receptor superfamily, member 8 | TRAF2 interacts with CD30. | BIND | 8662842 |
| TNFRSF9 | 4-1BB | CD137 | CDw137 | ILA | MGC2172 | tumor necrosis factor receptor superfamily, member 9 | - | HPRD,BioGRID | 9464265 |
| TNFSF14 | CD258 | HVEML | LIGHT | LTg | TR2 | tumor necrosis factor (ligand) superfamily, member 14 | Affinity Capture-MS Affinity Capture-Western | BioGRID | 12571250 |
| TNFSF4 | CD134L | CD252 | GP34 | OX-40L | OX4OL | TXGP1 | tumor necrosis factor (ligand) superfamily, member 4 | - | HPRD | 9418902 |
| TNFSF9 | 4-1BB-L | CD137L | tumor necrosis factor (ligand) superfamily, member 9 | - | HPRD,BioGRID | 9607925 |
| TNIK | - | TRAF2 and NCK interacting kinase | - | HPRD,BioGRID | 10521462 |
| TRADD | Hs.89862 | MGC11078 | TNFRSF1A-associated via death domain | - | HPRD,BioGRID | 8565075 |8702708 |10892748 |
| TRADD | Hs.89862 | MGC11078 | TNFRSF1A-associated via death domain | TRAF2 interacts with TRADD. This interaction was modeled on a demonstrated interaction between human TRAF2 and TRADD from an unspecified species. | BIND | 8702708 |
| TRADD | Hs.89862 | MGC11078 | TNFRSF1A-associated via death domain | TRADD interacts with TRAF2. This interaction was modeled on a demonstrated interaction between TRADD and TRAF2, both from an unspecified species. | BIND | 12796506 |
| TRAF1 | EBI6 | MGC:10353 | TNF receptor-associated factor 1 | TRAF2 interacts with TRAF1. This interaction was modeled on a demonstrated interaction between human TRAF2 and mouse TRAF1. | BIND | 8702708 |
| TRAF1 | EBI6 | MGC:10353 | TNF receptor-associated factor 1 | - | HPRD,BioGRID | 8702708 |
| TRAF2 | MGC:45012 | TRAP | TRAP3 | TNF receptor-associated factor 2 | - | HPRD,BioGRID | 8702708 |
| TRAF3IP2 | ACT1 | C6orf2 | C6orf4 | C6orf5 | C6orf6 | CIKS | DKFZp586G0522 | MGC3581 | TRAF3 interacting protein 2 | Two-hybrid | BioGRID | 16189514 |
| TRAF6 | MGC:3310 | RNF85 | TNF receptor-associated factor 6 | Two-hybrid | BioGRID | 16189514 |
| TRAIP | RNF206 | TRIP | TRAF interacting protein | - | HPRD,BioGRID | 9104814 |
| TRIM31 | C6orf13 | HCG1 | HCGI | RNF | tripartite motif-containing 31 | Two-hybrid | BioGRID | 16189514 |
| TRIM37 | KIAA0898 | MUL | POB1 | TEF3 | tripartite motif-containing 37 | - | HPRD,BioGRID | 11279055 |
| TRPC4AP | C20orf188 | TRRP4AP | TRUSS | transient receptor potential cation channel, subfamily C, member 4 associated protein | Affinity Capture-Western | BioGRID | 14585990 |
| TRPT1 | MGC11134 | tRNA phosphotransferase 1 | Two-hybrid | BioGRID | 16189514 |
| TTRAP | AD022 | EAP2 | MGC111021 | MGC9099 | dJ30M3.3 | TRAF and TNF receptor associated protein | - | HPRD,BioGRID | 10764746 |
| TUBA3C | TUBA2 | bA408E5.3 | tubulin, alpha 3c | - | HPRD | 14743216 |
| UBXN11 | COA-1 | DKFZp686F04228 | PP2243 | SOC | SOCI | UBXD5 | UBX domain protein 11 | Two-hybrid | BioGRID | 16189514 |
| USP2 | UBP41 | USP9 | ubiquitin specific peptidase 2 | Two-hybrid | BioGRID | 16189514 |
| USP53 | DKFZp781E1417 | ubiquitin specific peptidase 53 | Two-hybrid | BioGRID | 16189514 |
| USP7 | HAUSP | TEF1 | ubiquitin specific peptidase 7 (herpes virus-associated) | - | HPRD,BioGRID | 11279055 |
| ZBTB16 | PLZF | ZNF145 | zinc finger and BTB domain containing 16 | Two-hybrid | BioGRID | 16189514 |
| ZBTB25 | KUP | ZNF46 | zinc finger and BTB domain containing 25 | Two-hybrid | BioGRID | 16189514 |
| ZBTB43 | ZBTB22B | ZNF-X | ZNF297B | zinc finger and BTB domain containing 43 | Two-hybrid | BioGRID | 16189514 |
| ZFAND6 | AWP1 | ZA20D3 | ZFAND5B | zinc finger, AN1-type domain 6 | Two-hybrid | BioGRID | 16189514 |
| ZNF205 | ZNF210 | Zfp13 | zinc finger protein 205 | Two-hybrid | BioGRID | 16189514 |
| ZNF646 | KIAA0296 | zinc finger protein 646 | Two-hybrid | BioGRID | 16189514 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG MAPK SIGNALING PATHWAY | 267 | 205 | All SZGR 2.0 genes in this pathway |
| KEGG APOPTOSIS | 88 | 62 | All SZGR 2.0 genes in this pathway |
| KEGG RIG I LIKE RECEPTOR SIGNALING PATHWAY | 71 | 51 | All SZGR 2.0 genes in this pathway |
| KEGG ADIPOCYTOKINE SIGNALING PATHWAY | 67 | 57 | All SZGR 2.0 genes in this pathway |
| KEGG PATHWAYS IN CANCER | 328 | 259 | All SZGR 2.0 genes in this pathway |
| KEGG SMALL CELL LUNG CANCER | 84 | 67 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CERAMIDE PATHWAY | 22 | 16 | All SZGR 2.0 genes in this pathway |
| BIOCARTA HIVNEF PATHWAY | 58 | 43 | All SZGR 2.0 genes in this pathway |
| BIOCARTA DEATH PATHWAY | 33 | 24 | All SZGR 2.0 genes in this pathway |
| BIOCARTA KERATINOCYTE PATHWAY | 46 | 38 | All SZGR 2.0 genes in this pathway |
| BIOCARTA MAPK PATHWAY | 87 | 68 | All SZGR 2.0 genes in this pathway |
| BIOCARTA P38MAPK PATHWAY | 40 | 31 | All SZGR 2.0 genes in this pathway |
| BIOCARTA SODD PATHWAY | 10 | 7 | All SZGR 2.0 genes in this pathway |
| BIOCARTA TALL1 PATHWAY | 15 | 15 | All SZGR 2.0 genes in this pathway |
| BIOCARTA 41BB PATHWAY | 17 | 15 | All SZGR 2.0 genes in this pathway |
| BIOCARTA STRESS PATHWAY | 25 | 18 | All SZGR 2.0 genes in this pathway |
| BIOCARTA TNFR1 PATHWAY | 29 | 21 | All SZGR 2.0 genes in this pathway |
| BIOCARTA TNFR2 PATHWAY | 18 | 15 | All SZGR 2.0 genes in this pathway |
| ST TUMOR NECROSIS FACTOR PATHWAY | 29 | 20 | All SZGR 2.0 genes in this pathway |
| SIG CD40PATHWAYMAP | 34 | 28 | All SZGR 2.0 genes in this pathway |
| ST FAS SIGNALING PATHWAY | 65 | 54 | All SZGR 2.0 genes in this pathway |
| PID P38 MKK3 6PATHWAY | 26 | 21 | All SZGR 2.0 genes in this pathway |
| PID CD40 PATHWAY | 31 | 26 | All SZGR 2.0 genes in this pathway |
| PID TRAIL PATHWAY | 28 | 20 | All SZGR 2.0 genes in this pathway |
| PID TNF PATHWAY | 46 | 33 | All SZGR 2.0 genes in this pathway |
| PID CERAMIDE PATHWAY | 48 | 37 | All SZGR 2.0 genes in this pathway |
| PID HIV NEF PATHWAY | 35 | 26 | All SZGR 2.0 genes in this pathway |
| PID CASPASE PATHWAY | 52 | 39 | All SZGR 2.0 genes in this pathway |
| REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | 13 | 8 | All SZGR 2.0 genes in this pathway |
| REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | 30 | 26 | All SZGR 2.0 genes in this pathway |
| REACTOME TRAF6 MEDIATED NFKB ACTIVATION | 21 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME RIG I MDA5 MEDIATED INDUCTION OF IFN ALPHA BETA PATHWAYS | 73 | 57 | All SZGR 2.0 genes in this pathway |
| REACTOME APOPTOSIS | 148 | 94 | All SZGR 2.0 genes in this pathway |
| REACTOME INNATE IMMUNE SYSTEM | 279 | 178 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| DOANE RESPONSE TO ANDROGEN DN | 241 | 146 | All SZGR 2.0 genes in this pathway |
| GINESTIER BREAST CANCER ZNF217 AMPLIFIED DN | 335 | 193 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN DN | 770 | 415 | All SZGR 2.0 genes in this pathway |
| DARWICHE SKIN TUMOR PROMOTER UP | 142 | 96 | All SZGR 2.0 genes in this pathway |
| DARWICHE PAPILLOMA RISK LOW UP | 162 | 104 | All SZGR 2.0 genes in this pathway |
| DARWICHE PAPILLOMA RISK HIGH UP | 147 | 101 | All SZGR 2.0 genes in this pathway |
| DARWICHE SQUAMOUS CELL CARCINOMA DN | 181 | 107 | All SZGR 2.0 genes in this pathway |
| DARWICHE PAPILLOMA PROGRESSION RISK | 74 | 44 | All SZGR 2.0 genes in this pathway |
| DUTTA APOPTOSIS VIA NFKB | 33 | 25 | All SZGR 2.0 genes in this pathway |
| SCHEIDEREIT IKK INTERACTING PROTEINS | 58 | 45 | All SZGR 2.0 genes in this pathway |
| DARWICHE PAPILLOMA RISK HIGH VS LOW UP | 6 | 6 | All SZGR 2.0 genes in this pathway |
| YAGI AML SURVIVAL | 129 | 87 | All SZGR 2.0 genes in this pathway |
| ZHANG ANTIVIRAL RESPONSE TO RIBAVIRIN DN | 51 | 32 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL AND PROGENITOR | 681 | 420 | All SZGR 2.0 genes in this pathway |
| MONNIER POSTRADIATION TUMOR ESCAPE UP | 393 | 244 | All SZGR 2.0 genes in this pathway |
| DANG REGULATED BY MYC UP | 72 | 53 | All SZGR 2.0 genes in this pathway |
| ZEMBUTSU SENSITIVITY TO MITOMYCIN | 19 | 11 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE EARLY LATE | 317 | 190 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG RXRA BOUND 8D | 882 | 506 | All SZGR 2.0 genes in this pathway |