Gene Page: TRAF3
Summary ?
| GeneID | 7187 |
| Symbol | TRAF3 |
| Synonyms | CAP-1|CAP1|CD40bp|CRAF1|IIAE5|LAP1 |
| Description | TNF receptor associated factor 3 |
| Reference | MIM:601896|HGNC:HGNC:12033|Ensembl:ENSG00000131323|HPRD:03539|Vega:OTTHUMG00000171902 |
| Gene type | protein-coding |
| Map location | 14q32.32 |
| Pascal p-value | 0.035 |
| Fetal beta | -0.427 |
| eGene | Cerebellum Meta |
| Support | G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS G2Cdb.humanPSD G2Cdb.humanPSP |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWAScat | Genome-wide Association Studies | This data set includes 560 SNPs associated with schizophrenia. A total of 486 genes were mapped to these SNPs within 50kb. | |
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| Expression | Meta-analysis of gene expression | P value: 1.706 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
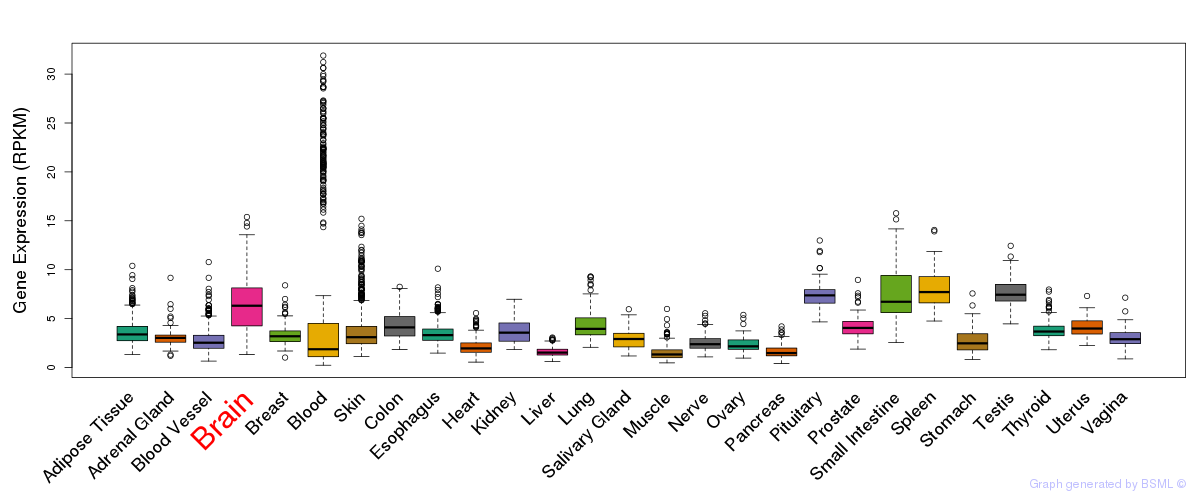
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004871 | signal transducer activity | TAS | 7530216 |7533327 | |
| GO:0005515 | protein binding | IPI | 7527023 |9162022 |10791955 |11035039 |12005438 |15280356 | |
| GO:0008270 | zinc ion binding | IEA | - | |
| GO:0046872 | metal ion binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006917 | induction of apoptosis | TAS | 10799510 | |
| GO:0007165 | signal transduction | TAS | 7530216 | |
| GO:0042981 | regulation of apoptosis | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005737 | cytoplasm | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ACTA1 | ACTA | ASMA | CFTD | CFTD1 | CFTDM | MPFD | NEM1 | NEM2 | NEM3 | actin, alpha 1, skeletal muscle | - | HPRD | 11950878 |
| CAP1 | CAP | CAP1-PEN | CAP, adenylate cyclase-associated protein 1 (yeast) | - | HPRD | 8761950 |
| CASP3 | CPP32 | CPP32B | SCA-1 | caspase 3, apoptosis-related cysteine peptidase | - | HPRD,BioGRID | 11261798 |
| CD27 | MGC20393 | S152 | T14 | TNFRSF7 | Tp55 | CD27 molecule | - | HPRD,BioGRID | 9582383 |9794406 |
| CD40 | Bp50 | CDW40 | MGC9013 | TNFRSF5 | p50 | CD40 molecule, TNF receptor superfamily member 5 | - | HPRD | 7527023 |7530216 |7533327 |10984535 |
| CD40 | Bp50 | CDW40 | MGC9013 | TNFRSF5 | p50 | CD40 molecule, TNF receptor superfamily member 5 | Affinity Capture-Western Co-crystal Structure Reconstituted Complex Two-hybrid | BioGRID | 7527023 |9384571 |9990007 |10984535 |
| CD40 | Bp50 | CDW40 | MGC9013 | TNFRSF5 | p50 | CD40 molecule, TNF receptor superfamily member 5 | CD40 interacts with TRAF3. | BIND | 10352240 |
| CD40 | Bp50 | CDW40 | MGC9013 | TNFRSF5 | p50 | CD40 molecule, TNF receptor superfamily member 5 | - | HPRD | 7527023 |7533327 |10984535 |
| CFLAR | CASH | CASP8AP1 | CLARP | Casper | FLAME | FLAME-1 | FLAME1 | FLIP | I-FLICE | MRIT | c-FLIP | c-FLIPL | c-FLIPR | c-FLIPS | CASP8 and FADD-like apoptosis regulator | Affinity Capture-Western | BioGRID | 10837247 |
| EDA2R | EDA-A2R | EDAA2R | TNFRSF27 | XEDAR | ectodysplasin A2 receptor | Affinity Capture-Western | BioGRID | 12270937 |
| GRIN1 | NMDA1 | NMDAR1 | NR1 | glutamate receptor, ionotropic, N-methyl D-aspartate 1 | - | HPRD | 10862698 |
| LTBR | CD18 | D12S370 | LT-BETA-R | TNF-R-III | TNFCR | TNFR-RP | TNFR2-RP | TNFRSF3 | lymphotoxin beta receptor (TNFR superfamily, member 3) | - | HPRD,BioGRID | 9122217 |
| LTBR | CD18 | D12S370 | LT-BETA-R | TNF-R-III | TNFCR | TNFR-RP | TNFR2-RP | TNFRSF3 | lymphotoxin beta receptor (TNFR superfamily, member 3) | TRAF3 interacts with LTBR. | BIND | 12571250 |
| MAP2K1 | MAPKK1 | MEK1 | MKK1 | PRKMK1 | mitogen-activated protein kinase kinase 1 | - | HPRD | 8626767 |
| MAP3K14 | FTDCR1B | HS | HSNIK | NIK | mitogen-activated protein kinase kinase kinase 14 | - | HPRD,BioGRID | 11278268 |
| MAP3K14 | FTDCR1B | HS | HSNIK | NIK | mitogen-activated protein kinase kinase kinase 14 | NIK interacts with TRAF3. This interaction was modeled on a demonstrated interaction between human NIK and TRAF3 from an unspecified species. | BIND | 9275204 |
| MAP3K5 | ASK1 | MAPKKK5 | MEKK5 | mitogen-activated protein kinase kinase kinase 5 | Affinity Capture-Western | BioGRID | 9774977 |
| NGFR | CD271 | Gp80-LNGFR | TNFRSF16 | p75(NTR) | p75NTR | nerve growth factor receptor (TNFR superfamily, member 16) | Affinity Capture-Western | BioGRID | 10514511 |
| NUP62 | DKFZp547L134 | FLJ20822 | FLJ43869 | IBSN | MGC841 | SNDI | p62 | nucleoporin 62kDa | in vitro in vivo Two-hybrid | BioGRID | 10781837 |
| RIPK1 | FLJ39204 | RIP | RIP1 | receptor (TNFRSF)-interacting serine-threonine kinase 1 | - | HPRD,BioGRID | 8612133 |
| RRAS2 | TC21 | related RAS viral (r-ras) oncogene homolog 2 | - | HPRD | 10557073 |
| SRC | ASV | SRC1 | c-SRC | p60-Src | v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (avian) | Affinity Capture-Western | BioGRID | 10635328 |
| TANK | I-TRAF | TRAF2 | TRAF family member-associated NFKB activator | - | HPRD,BioGRID | 8710854 |
| TNFRSF11A | CD265 | FEO | LOH18CR1 | ODFR | OFE | OPTB7 | OSTS | PDB2 | RANK | TRANCER | tumor necrosis factor receptor superfamily, member 11a, NFKB activator | - | HPRD,BioGRID | 9774460 |9852070 |10025951 |
| TNFRSF12A | CD266 | FN14 | TWEAKR | tumor necrosis factor receptor superfamily, member 12A | - | HPRD | 11728344 |
| TNFRSF14 | ATAR | HVEA | HVEM | LIGHTR | TR2 | tumor necrosis factor receptor superfamily, member 14 (herpesvirus entry mediator) | - | HPRD,BioGRID | 9162022 |
| TNFRSF17 | BCM | BCMA | CD269 | tumor necrosis factor receptor superfamily, member 17 | - | HPRD,BioGRID | 10903733 |
| TNFRSF18 | AITR | GITR | GITR-D | tumor necrosis factor receptor superfamily, member 18 | - | HPRD,BioGRID | 10037686 |
| TNFRSF19 | TAJ | TAJ-alpha | TRADE | TROY | tumor necrosis factor receptor superfamily, member 19 | - | HPRD,BioGRID | 10809768 |
| TNFRSF4 | ACT35 | CD134 | OX40 | TXGP1L | tumor necrosis factor receptor superfamily, member 4 | - | HPRD | 9418902 |
| TNFRSF8 | CD30 | D1S166E | KI-1 | tumor necrosis factor receptor superfamily, member 8 | CRAF1 interacts with CD30. | BIND | 8662842 |
| TNFRSF8 | CD30 | D1S166E | KI-1 | tumor necrosis factor receptor superfamily, member 8 | - | HPRD,BioGRID | 8943059 |
| TNFRSF9 | 4-1BB | CD137 | CDw137 | ILA | MGC2172 | tumor necrosis factor receptor superfamily, member 9 | - | HPRD,BioGRID | 9464265 |
| TNFSF14 | CD258 | HVEML | LIGHT | LTg | TR2 | tumor necrosis factor (ligand) superfamily, member 14 | Affinity Capture-MS Affinity Capture-Western | BioGRID | 12571250 |
| TRADD | Hs.89862 | MGC11078 | TNFRSF1A-associated via death domain | - | HPRD | 8565075 |
| TRAF3IP1 | DKFZp434F124 | MIP-T3 | MIPT3 | TNF receptor-associated factor 3 interacting protein 1 | - | HPRD,BioGRID | 10791955 |
| TRAF3IP2 | ACT1 | C6orf2 | C6orf4 | C6orf5 | C6orf6 | CIKS | DKFZp586G0522 | MGC3581 | TRAF3 interacting protein 2 | - | HPRD,BioGRID | 12089335 |
| TRAF3IP3 | DJ434O14.3 | FLJ44151 | MGC117354 | MGC163289 | T3JAM | TRAF3 interacting protein 3 | - | HPRD | 14572659 |
| TRIM37 | KIAA0898 | MUL | POB1 | TEF3 | tripartite motif-containing 37 | - | HPRD,BioGRID | 11279055 |
| TTRAP | AD022 | EAP2 | MGC111021 | MGC9099 | dJ30M3.3 | TRAF and TNF receptor associated protein | Affinity Capture-Western | BioGRID | 10764746 |
| USP7 | HAUSP | TEF1 | ubiquitin specific peptidase 7 (herpes virus-associated) | - | HPRD,BioGRID | 11279055 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG TOLL LIKE RECEPTOR SIGNALING PATHWAY | 102 | 88 | All SZGR 2.0 genes in this pathway |
| KEGG RIG I LIKE RECEPTOR SIGNALING PATHWAY | 71 | 51 | All SZGR 2.0 genes in this pathway |
| KEGG PATHWAYS IN CANCER | 328 | 259 | All SZGR 2.0 genes in this pathway |
| KEGG SMALL CELL LUNG CANCER | 84 | 67 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CD40 PATHWAY | 15 | 13 | All SZGR 2.0 genes in this pathway |
| BIOCARTA TALL1 PATHWAY | 15 | 15 | All SZGR 2.0 genes in this pathway |
| BIOCARTA TNFR2 PATHWAY | 18 | 15 | All SZGR 2.0 genes in this pathway |
| ST ERK1 ERK2 MAPK PATHWAY | 32 | 25 | All SZGR 2.0 genes in this pathway |
| SIG CD40PATHWAYMAP | 34 | 28 | All SZGR 2.0 genes in this pathway |
| PID CD40 PATHWAY | 31 | 26 | All SZGR 2.0 genes in this pathway |
| REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | 14 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | 14 | 7 | All SZGR 2.0 genes in this pathway |
| REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | 31 | 19 | All SZGR 2.0 genes in this pathway |
| REACTOME RIG I MDA5 MEDIATED INDUCTION OF IFN ALPHA BETA PATHWAYS | 73 | 57 | All SZGR 2.0 genes in this pathway |
| REACTOME INNATE IMMUNE SYSTEM | 279 | 178 | All SZGR 2.0 genes in this pathway |
| REACTOME ACTIVATED TLR4 SIGNALLING | 93 | 69 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME TOLL RECEPTOR CASCADES | 118 | 84 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA UP | 783 | 507 | All SZGR 2.0 genes in this pathway |
| BERTUCCI MEDULLARY VS DUCTAL BREAST CANCER UP | 206 | 111 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS MESENCHYMAL DN | 460 | 312 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER BASAL VS MESENCHYMAL DN | 50 | 36 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION UP | 1278 | 748 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS UP | 214 | 155 | All SZGR 2.0 genes in this pathway |
| LAU APOPTOSIS CDKN2A UP | 55 | 40 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA UP | 536 | 340 | All SZGR 2.0 genes in this pathway |
| RASHI RESPONSE TO IONIZING RADIATION 2 | 127 | 92 | All SZGR 2.0 genes in this pathway |
| GOTZMANN EPITHELIAL TO MESENCHYMAL TRANSITION UP | 69 | 55 | All SZGR 2.0 genes in this pathway |
| SHIN B CELL LYMPHOMA CLUSTER 2 | 30 | 23 | All SZGR 2.0 genes in this pathway |
| MORI PRE BI LYMPHOCYTE DN | 77 | 49 | All SZGR 2.0 genes in this pathway |
| MORI IMMATURE B LYMPHOCYTE UP | 53 | 35 | All SZGR 2.0 genes in this pathway |
| OKUMURA INFLAMMATORY RESPONSE LPS | 183 | 115 | All SZGR 2.0 genes in this pathway |
| THEILGAARD NEUTROPHIL AT SKIN WOUND UP | 77 | 52 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 16HR UP | 225 | 139 | All SZGR 2.0 genes in this pathway |
| ZHANG RESPONSE TO CANTHARIDIN UP | 19 | 12 | All SZGR 2.0 genes in this pathway |
| NATSUME RESPONSE TO INTERFERON BETA DN | 52 | 33 | All SZGR 2.0 genes in this pathway |
| MURAKAMI UV RESPONSE 6HR UP | 37 | 31 | All SZGR 2.0 genes in this pathway |
| JACKSON DNMT1 TARGETS UP | 77 | 57 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 2 | 473 | 224 | All SZGR 2.0 genes in this pathway |
| FOSTER TOLERANT MACROPHAGE UP | 156 | 92 | All SZGR 2.0 genes in this pathway |
| MITSIADES RESPONSE TO APLIDIN DN | 249 | 165 | All SZGR 2.0 genes in this pathway |
| BONOME OVARIAN CANCER SURVIVAL SUBOPTIMAL DEBULKING | 510 | 309 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO CSF2RB AND IL4 UP | 338 | 225 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF UP | 418 | 282 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF VS CSF2RB AND IL4 UP | 408 | 276 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER SUBCLASS S1 | 237 | 159 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| TORCHIA TARGETS OF EWSR1 FLI1 FUSION DN | 321 | 200 | All SZGR 2.0 genes in this pathway |
| KRIEG HYPOXIA NOT VIA KDM3A | 770 | 480 | All SZGR 2.0 genes in this pathway |
| PHONG TNF RESPONSE NOT VIA P38 | 337 | 236 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-25/32/92/363/367 | 461 | 467 | m8 | hsa-miR-25brain | CAUUGCACUUGUCUCGGUCUGA |
| hsa-miR-32 | UAUUGCACAUUACUAAGUUGC | ||||
| hsa-miR-92 | UAUUGCACUUGUCCCGGCCUG | ||||
| hsa-miR-367 | AAUUGCACUUUAGCAAUGGUGA | ||||
| hsa-miR-92bSZ | UAUUGCACUCGUCCCGGCCUC | ||||
| miR-494 | 470 | 476 | 1A | hsa-miR-494brain | UGAAACAUACACGGGAAACCUCUU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.