Gene Page: UBE2I
Summary ?
| GeneID | 7329 |
| Symbol | UBE2I |
| Synonyms | C358B7.1|P18|UBC9 |
| Description | ubiquitin conjugating enzyme E2 I |
| Reference | MIM:601661|HGNC:HGNC:12485|Ensembl:ENSG00000103275|HPRD:09045|Vega:OTTHUMG00000186701 |
| Gene type | protein-coding |
| Map location | 16p13.3 |
| Pascal p-value | 0.05 |
| Sherlock p-value | 0.393 |
| Fetal beta | 0.751 |
| DMG | 1 (# studies) |
| eGene | Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0725 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg07982263 | 16 | 1359166 | UBE2I | 9.53E-8 | -0.006 | 2.14E-5 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
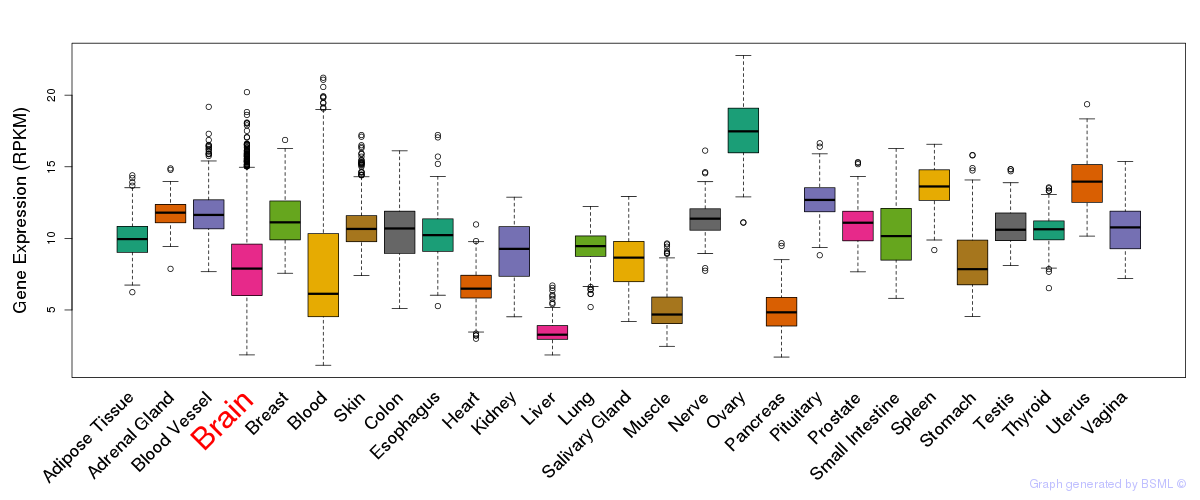
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| TRA2B | 0.92 | 0.89 |
| DKC1 | 0.91 | 0.90 |
| SFRS3 | 0.91 | 0.89 |
| EIF4A3 | 0.90 | 0.90 |
| MED17 | 0.90 | 0.91 |
| MCM6 | 0.90 | 0.91 |
| UBE2I | 0.90 | 0.90 |
| NUP107 | 0.89 | 0.91 |
| ZIK1 | 0.89 | 0.88 |
| SFRS7 | 0.89 | 0.89 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| HLA-F | -0.69 | -0.76 |
| MT-CO2 | -0.68 | -0.81 |
| AF347015.33 | -0.68 | -0.81 |
| AF347015.27 | -0.68 | -0.80 |
| AF347015.31 | -0.67 | -0.80 |
| MT-CYB | -0.66 | -0.79 |
| TINAGL1 | -0.66 | -0.75 |
| AIFM3 | -0.65 | -0.72 |
| AF347015.8 | -0.65 | -0.80 |
| PTH1R | -0.65 | -0.74 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005515 | protein binding | IPI | 9885291 |12072434 |14516784 | |
| GO:0016874 | ligase activity | IEA | - | |
| GO:0019787 | small conjugating protein ligase activity | IEA | - | |
| GO:0043398 | HLH domain binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006511 | ubiquitin-dependent protein catabolic process | IEA | - | |
| GO:0006511 | ubiquitin-dependent protein catabolic process | TAS | 8565643 | |
| GO:0007049 | cell cycle | IEA | - | |
| GO:0007067 | mitosis | IEA | - | |
| GO:0007059 | chromosome segregation | IEA | - | |
| GO:0051246 | regulation of protein metabolic process | IEA | - | |
| GO:0051301 | cell division | IEA | - | |
| GO:0043687 | post-translational protein modification | IEA | - | |
| GO:0044419 | interspecies interaction between organisms | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000795 | synaptonemal complex | TAS | 8610150 | |
| GO:0005634 | nucleus | IEA | - | |
| GO:0016605 | PML body | IDA | 9885291 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| APEX1 | APE | APE-1 | APE1 | APEN | APEX | APX | HAP1 | REF-1 | REF1 | APEX nuclease (multifunctional DNA repair enzyme) 1 | - | HPRD,BioGRID | 12110916 |
| AR | AIS | DHTR | HUMARA | KD | NR3C4 | SBMA | SMAX1 | TFM | androgen receptor | Affinity Capture-Western Phenotypic Enhancement Two-hybrid | BioGRID | 10383460 |
| AR | AIS | DHTR | HUMARA | KD | NR3C4 | SBMA | SMAX1 | TFM | androgen receptor | - | HPRD | 8798622 |
| ATF2 | CRE-BP1 | CREB2 | HB16 | MGC111558 | TREB7 | activating transcription factor 2 | - | HPRD,BioGRID | 9488727 |
| BLMH | BH | BMH | bleomycin hydrolase | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 9855622 |
| C18orf25 | ARKL1 | MGC12909 | MGC87799 | chromosome 18 open reading frame 25 | Two-hybrid | BioGRID | 16189514 |
| CBX4 | NBP16 | PC2 | hPC2 | chromobox homolog 4 (Pc class homolog, Drosophila) | Ubc9 interacts with Pc2. | BIND | 15592428 |
| CEBPE | C/EBP-epsilon | CRP1 | CCAAT/enhancer binding protein (C/EBP), epsilon | C/EBP-epsilon interacts with UBE2I. | BIND | 15588942 |
| CHD3 | Mi-2a | Mi2-ALPHA | ZFH | chromodomain helicase DNA binding protein 3 | - | HPRD,BioGRID | 14609633 |
| CHD3 | Mi-2a | Mi2-ALPHA | ZFH | chromodomain helicase DNA binding protein 3 | Ubc9 interacts with CHD3. | BIND | 14609633 |
| CHMP4B | C20orf178 | CHMP4A | CTPP3 | SNF7 | SNF7-2 | Shax1 | dJ553F4.4 | chromatin modifying protein 4B | Two-hybrid | BioGRID | 16189514 |
| CHUK | IKBKA | IKK-alpha | IKK1 | IKKA | NFKBIKA | TCF16 | conserved helix-loop-helix ubiquitous kinase | Biochemical Activity | BioGRID | 11853669 |
| DACH1 | DACH | FLJ10138 | dachshund homolog 1 (Drosophila) | - | HPRD,BioGRID | 11025202 |
| DAXX | BING2 | DAP6 | EAP1 | MGC126245 | MGC126246 | death-domain associated protein | Daxx interacts with Ubc9. | BIND | 11112409 |
| DAXX | BING2 | DAP6 | EAP1 | MGC126245 | MGC126246 | death-domain associated protein | Biochemical Activity Two-hybrid | BioGRID | 11112409 |18691969 |
| DNMT3A | DNMT3A2 | M.HsaIIIA | DNA (cytosine-5-)-methyltransferase 3 alpha | - | HPRD,BioGRID | 14752048 |
| DNMT3B | ICF | M.HsaIIIB | DNA (cytosine-5-)-methyltransferase 3 beta | Dnmt3b interacts with Ubc9. | BIND | 11735126 |
| DNMT3B | ICF | M.HsaIIIB | DNA (cytosine-5-)-methyltransferase 3 beta | - | HPRD,BioGRID | 11735126 |
| EP300 | KAT3B | p300 | E1A binding protein p300 | - | HPRD | 8824223 |
| ESR1 | DKFZp686N23123 | ER | ESR | ESRA | Era | NR3A1 | estrogen receptor 1 | ER-alpha interacts with an unspecified isoform of Ubc9. | BIND | 15666801 |
| ETS1 | ETS-1 | EWSR2 | FLJ10768 | v-ets erythroblastosis virus E26 oncogene homolog 1 (avian) | - | HPRD,BioGRID | 9333025 |
| ETV6 | TEL | TEL/ABL | ets variant 6 | - | HPRD | 10377438 |
| FAS | ALPS1A | APO-1 | APT1 | CD95 | FAS1 | FASTM | TNFRSF6 | Fas (TNF receptor superfamily, member 6) | - | HPRD,BioGRID | 9257699 |
| FHIT | AP3Aase | FRA3B | fragile histidine triad gene | - | HPRD,BioGRID | 11085938 |
| FMR1 | FMRP | FRAXA | MGC87458 | POF | POF1 | fragile X mental retardation 1 | - | HPRD,BioGRID | 11367701 |
| HDAC4 | HA6116 | HD4 | HDAC-A | HDACA | KIAA0288 | histone deacetylase 4 | Biochemical Activity | BioGRID | 18691969 |
| HGS | HRS | ZFYVE8 | hepatocyte growth factor-regulated tyrosine kinase substrate | Two-hybrid | BioGRID | 16189514 |
| HIPK2 | DKFZp686K02111 | FLJ23711 | PRO0593 | homeodomain interacting protein kinase 2 | Ubc9 with HIPK2. | BIND | 14609633 |
| HIPK2 | DKFZp686K02111 | FLJ23711 | PRO0593 | homeodomain interacting protein kinase 2 | Two-hybrid | BioGRID | 14609633 |
| HIPK2 | DKFZp686K02111 | FLJ23711 | PRO0593 | homeodomain interacting protein kinase 2 | - | HPRD | 10535925 |
| HNRNPC | C1 | C2 | HNRNP | HNRPC | MGC104306 | MGC105117 | MGC117353 | MGC131677 | SNRPC | heterogeneous nuclear ribonucleoprotein C (C1/C2) | Two-hybrid | BioGRID | 16189514 |
| HNRNPK | CSBP | FLJ41122 | HNRPK | TUNP | heterogeneous nuclear ribonucleoprotein K | Two-hybrid | BioGRID | 16189514 |
| HSF2 | MGC117376 | MGC156196 | MGC75048 | heat shock transcription factor 2 | Two-hybrid | BioGRID | 11278381 |
| IKZF1 | Hs.54452 | IK1 | IKAROS | LYF1 | PRO0758 | ZNFN1A1 | hIk-1 | IKAROS family zinc finger 1 (Ikaros) | Two-hybrid | BioGRID | 16189514 |
| IKZF3 | AIO | AIOLOS | ZNFN1A3 | IKAROS family zinc finger 3 (Aiolos) | Two-hybrid | BioGRID | 16189514 |
| IPO13 | IMP13 | KAP13 | KIAA0724 | RANBP13 | importin 13 | - | HPRD,BioGRID | 11447110 |
| JUN | AP-1 | AP1 | c-Jun | jun oncogene | - | HPRD,BioGRID | 8733011 |
| KCNK1 | DPK | HOHO | K2p1.1 | KCNO1 | TWIK-1 | TWIK1 | potassium channel, subfamily K, member 1 | K2P1 interacts with Ubc-9. | BIND | 15820677 |
| MAP3K1 | MAPKKK1 | MEKK | MEKK1 | mitogen-activated protein kinase kinase kinase 1 | Reconstituted Complex Two-hybrid | BioGRID | 9563508 |
| MAPK1IP1L | C14orf32 | MGC23138 | MISS | c14_5346 | mitogen-activated protein kinase 1 interacting protein 1-like | Two-hybrid | BioGRID | 16189514 |
| MITF | MI | WS2A | bHLHe32 | microphthalmia-associated transcription factor | - | HPRD,BioGRID | 10694430 |
| MYB | Cmyb | c-myb | c-myb_CDS | efg | v-myb myeloblastosis viral oncogene homolog (avian) | - | HPRD | 11779867 |
| NR3C1 | GCCR | GCR | GR | GRL | nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) | - | HPRD | 11812797 |
| NUDCD3 | KIAA1068 | NudCL | NudC domain containing 3 | Two-hybrid | BioGRID | 16189514 |
| PDLIM7 | LMP1 | PDZ and LIM domain 7 (enigma) | Two-hybrid | BioGRID | 16189514 |
| PDZK1 | CAP70 | CLAMP | NHERF3 | PDZD1 | PDZ domain containing 1 | Two-hybrid | BioGRID | 14531806 |
| PHC1 | EDR1 | HPH1 | RAE28 | polyhomeotic homolog 1 (Drosophila) | Two-hybrid | BioGRID | 16189514 |
| PIAS1 | DDXBP1 | GBP | GU/RH-II | MGC141878 | MGC141879 | ZMIZ3 | protein inhibitor of activated STAT, 1 | Ubc9 interacts with PIAS1. | BIND | 14609633 |
| PIAS1 | DDXBP1 | GBP | GU/RH-II | MGC141878 | MGC141879 | ZMIZ3 | protein inhibitor of activated STAT, 1 | - | HPRD | 12177000 |
| PIAS1 | DDXBP1 | GBP | GU/RH-II | MGC141878 | MGC141879 | ZMIZ3 | protein inhibitor of activated STAT, 1 | Reconstituted Complex Two-hybrid | BioGRID | 11583632 |12356736 |14609633 |
| PIAS2 | MGC102682 | MIZ1 | PIASX | PIASX-ALPHA | PIASX-BETA | SIZ2 | ZMIZ4 | miz | protein inhibitor of activated STAT, 2 | Two-hybrid | BioGRID | 14609633 |16189514 |
| PIAS2 | MGC102682 | MIZ1 | PIASX | PIASX-ALPHA | PIASX-BETA | SIZ2 | ZMIZ4 | miz | protein inhibitor of activated STAT, 2 | Ubc9 interacts with PIASxb-eta. | BIND | 14609633 |
| PML | MYL | PP8675 | RNF71 | TRIM19 | promyelocytic leukemia | Biochemical Activity | BioGRID | 18691969 |
| PPM1J | DKFZp434P1514 | FLJ35951 | MGC19531 | MGC90149 | PP2CZ | PP2Czeta | PPP2CZ | protein phosphatase 1J (PP2C domain containing) | - | HPRD,BioGRID | 12633878 |
| PTEN | 10q23del | BZS | MGC11227 | MHAM | MMAC1 | PTEN1 | TEP1 | phosphatase and tensin homolog | Affinity Capture-Western | BioGRID | 12620973 |
| RAD51 | BRCC5 | HRAD51 | HsRad51 | HsT16930 | RAD51A | RECA | RAD51 homolog (RecA homolog, E. coli) (S. cerevisiae) | - | HPRD,BioGRID | 8610150 |
| RAD52 | - | RAD52 homolog (S. cerevisiae) | Two-hybrid | BioGRID | 8921390 |
| RAD54B | FSBP | RDH54 | RAD54 homolog B (S. cerevisiae) | Two-hybrid | BioGRID | 16189514 |
| RANBP2 | NUP358 | TRP1 | TRP2 | RAN binding protein 2 | Ubc9 interacts with RanBP2. | BIND | 15608651 |
| RANBP2 | NUP358 | TRP1 | TRP2 | RAN binding protein 2 | RanBP2 interacts with Ubc9. | BIND | 15378033 |
| RANBP2 | NUP358 | TRP1 | TRP2 | RAN binding protein 2 | - | HPRD,BioGRID | 12192048 |
| RANGAP1 | Fug1 | KIAA1835 | MGC20266 | SD | Ran GTPase activating protein 1 | Affinity Capture-MS Biochemical Activity Reconstituted Complex | BioGRID | 12924945 |17353931 |18691969 |
| RANGAP1 | Fug1 | KIAA1835 | MGC20266 | SD | Ran GTPase activating protein 1 | Unspecified isoform of UBC9 conjugated with SUMO-1 sumoylates RanGAP1. This interaction was modeled on a demonstrated interaction between UBC9 from an unspecified species and RanGAP1 from an unspecified species. | BIND | 15546615 |
| RANGAP1 | Fug1 | KIAA1835 | MGC20266 | SD | Ran GTPase activating protein 1 | - | HPRD | 11853669 |
| RB1 | OSRC | RB | p105-Rb | pRb | pp110 | retinoblastoma 1 | RB1 (pRb) interacts with, and is sumoylated by, an unspecified isoform of UBE2I (Ubc9). | BIND | 15806172 |
| RIPK2 | CARD3 | CARDIAK | CCK | GIG30 | RICK | RIP2 | receptor-interacting serine-threonine kinase 2 | Two-hybrid | BioGRID | 16189514 |
| SAE1 | AOS1 | FLJ3091 | HSPC140 | SUA1 | SUMO1 activating enzyme subunit 1 | Affinity Capture-MS | BioGRID | 17353931 |
| SALL1 | HSAL1 | TBS | ZNF794 | sal-like 1 (Drosophila) | SALL1 interacts with UBE2I. | BIND | 12200128 |
| SALL1 | HSAL1 | TBS | ZNF794 | sal-like 1 (Drosophila) | - | HPRD,BioGRID | 12200128 |
| SATB1 | - | SATB homeobox 1 | Two-hybrid | BioGRID | 16189514 |
| SCNN1A | ENaCa | ENaCalpha | FLJ21883 | SCNEA | SCNN1 | sodium channel, nonvoltage-gated 1 alpha | - | HPRD,BioGRID | 14556380 |
| SIAH1 | FLJ08065 | HUMSIAH | Siah-1 | Siah-1a | hSIAH1 | seven in absentia homolog 1 (Drosophila) | Ubc9 interacts with Siah-1. | BIND | 9334332 |
| SIAH1 | FLJ08065 | HUMSIAH | Siah-1 | Siah-1a | hSIAH1 | seven in absentia homolog 1 (Drosophila) | hSiah1 interacts with Ubc9. | BIND | 9334332 |
| SIAH2 | hSiah2 | seven in absentia homolog 2 (Drosophila) | hSiah2 interacts with Ubc9. | BIND | 9334332 |
| SIAH2 | hSiah2 | seven in absentia homolog 2 (Drosophila) | Ubc9 interacts with Siah-2. | BIND | 9334332 |
| SLC2A1 | DYT17 | DYT18 | GLUT | GLUT1 | MGC141895 | MGC141896 | PED | solute carrier family 2 (facilitated glucose transporter), member 1 | - | HPRD | 11842083 |
| SLC2A4 | GLUT4 | solute carrier family 2 (facilitated glucose transporter), member 4 | - | HPRD | 11842083 |
| SMAD4 | DPC4 | JIP | MADH4 | SMAD family member 4 | Two-hybrid | BioGRID | 16189514 |
| SP100 | DKFZp686E07254 | FLJ00340 | FLJ34579 | SP100 nuclear antigen | Biochemical Activity Reconstituted Complex | BioGRID | 18691969 |
| SREBF1 | SREBP-1c | SREBP1 | bHLHd1 | sterol regulatory element binding transcription factor 1 | - | HPRD,BioGRID | 12615929 |
| SREBF2 | SREBP2 | bHLHd2 | sterol regulatory element binding transcription factor 2 | - | HPRD,BioGRID | 12615929 |
| STAT1 | DKFZp686B04100 | ISGF-3 | STAT91 | signal transducer and activator of transcription 1, 91kDa | - | HPRD | 12356736 |
| SUMO1 | DAP-1 | GMP1 | OFC10 | PIC1 | SENP2 | SMT3 | SMT3C | SMT3H3 | SUMO-1 | UBL1 | SMT3 suppressor of mif two 3 homolog 1 (S. cerevisiae) | - | HPRD,BioGRID | 12924945 |
| SUMO1 | DAP-1 | GMP1 | OFC10 | PIC1 | SENP2 | SMT3 | SMT3C | SMT3H3 | SUMO-1 | UBL1 | SMT3 suppressor of mif two 3 homolog 1 (S. cerevisiae) | Ubc9 interacts with SUMO1. | BIND | 15592428 |
| SUMO1 | DAP-1 | GMP1 | OFC10 | PIC1 | SENP2 | SMT3 | SMT3C | SMT3H3 | SUMO-1 | UBL1 | SMT3 suppressor of mif two 3 homolog 1 (S. cerevisiae) | hUbc9 interacts with SUMO-1. | BIND | 9920803 |
| SUMO2 | HSMT3 | MGC117191 | SMT3B | SMT3H2 | SMT3 suppressor of mif two 3 homolog 2 (S. cerevisiae) | - | HPRD | 12924945 |
| SUMO3 | SMT3A | SMT3H1 | SUMO-3 | SMT3 suppressor of mif two 3 homolog 3 (S. cerevisiae) | Affinity Capture-MS | BioGRID | 17353931 |
| SUMO3 | SMT3A | SMT3H1 | SUMO-3 | SMT3 suppressor of mif two 3 homolog 3 (S. cerevisiae) | - | HPRD | 12924945 |
| TCF3 | E2A | ITF1 | MGC129647 | MGC129648 | bHLHb21 | transcription factor 3 (E2A immunoglobulin enhancer binding factors E12/E47) | Reconstituted Complex Two-hybrid | BioGRID | 10497239 |
| TDG | - | thymine-DNA glycosylase | Biochemical Activity | BioGRID | 18691969 |
| TNFRSF1A | CD120a | FPF | MGC19588 | TBP1 | TNF-R | TNF-R-I | TNF-R55 | TNFAR | TNFR1 | TNFR55 | TNFR60 | p55 | p55-R | p60 | tumor necrosis factor receptor superfamily, member 1A | - | HPRD,BioGRID | 9563508 |
| TOP1 | TOPI | topoisomerase (DNA) I | - | HPRD,BioGRID | 10759568 |
| TOPORS | LUN | P53BP3 | RP31 | TP53BPL | topoisomerase I binding, arginine/serine-rich | - | HPRD,BioGRID | 14516784 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | - | HPRD,BioGRID | 8921390 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | p53 interacts with Ubc9. This interaction was modelled on a demonstrated interaction between p53 from human and Ubc9 from an unspecified species. | BIND | 10788439 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | UBE2I interacts with TP53. This interaction was modelled on a demonstrated interaction between D. melanogaster Lwr (DmUbc9) and p53 from an unspecified species. | BIND | 9514881 |
| TP73 | P73 | tumor protein p73 | Two-hybrid | BioGRID | 10961991 |
| TRAF4 | CART1 | MLN62 | RNF83 | TNF receptor-associated factor 4 | TRAF4 interacts with UBC9. | BIND | 12023963 |
| TRIM29 | ATDC | FLJ36085 | tripartite motif-containing 29 | Two-hybrid | BioGRID | 16189514 |
| TRIM54 | MURF | MURF-3 | RNF30 | tripartite motif-containing 54 | Two-hybrid | BioGRID | 11927605 |
| TRIM55 | MURF-2 | RNF29 | tripartite motif-containing 55 | Two-hybrid | BioGRID | 11927605 |
| TRIM63 | FLJ32380 | IRF | MURF1 | MURF2 | RNF28 | SMRZ | tripartite motif-containing 63 | Two-hybrid | BioGRID | 11927605 |
| TTRAP | AD022 | EAP2 | MGC111021 | MGC9099 | dJ30M3.3 | TRAF and TNF receptor associated protein | Two-hybrid | BioGRID | 14609633 |
| TTRAP | AD022 | EAP2 | MGC111021 | MGC9099 | dJ30M3.3 | TRAF and TNF receptor associated protein | Ubc9 interacts with TTRAP. | BIND | 14609633 |
| UBA2 | ARX | FLJ13058 | HRIHFB2115 | SAE2 | ubiquitin-like modifier activating enzyme 2 | Affinity Capture-MS Biochemical Activity | BioGRID | 17353931 |18691969 |
| UBE2I | C358B7.1 | P18 | UBC9 | ubiquitin-conjugating enzyme E2I (UBC9 homolog, yeast) | Two-hybrid | BioGRID | 8610150 |
| UBE2K | DKFZp564C1216 | DKFZp686J24237 | E2-25K | HIP2 | HYPG | LIG | ubiquitin-conjugating enzyme E2K (UBC1 homolog, yeast) | Biochemical Activity | BioGRID | 18691969 |
| UCHL1 | PARK5 | PGP9.5 | Uch-L1 | ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase) | Two-hybrid | BioGRID | 12082530 |
| UNC119 | HRG4 | unc-119 homolog (C. elegans) | Two-hybrid | BioGRID | 16169070 |
| WT1 | GUD | WAGR | WIT-2 | WT33 | Wilms tumor 1 | - | HPRD,BioGRID | 8798754 |
| ZBED1 | ALTE | DREF | KIAA0785 | TRAMP | hDREF | zinc finger, BED-type containing 1 | Two-hybrid | BioGRID | 16189514 |
| ZNF451 | COASTER | FLJ23421 | FLJ53246 | FLJ90693 | KIAA0576 | KIAA1702 | MGC26701 | dJ417I1.1 | zinc finger protein 451 | Two-hybrid | BioGRID | 16189514 |
| ZNF646 | KIAA0296 | zinc finger protein 646 | Two-hybrid | BioGRID | 16189514 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG UBIQUITIN MEDIATED PROTEOLYSIS | 138 | 98 | All SZGR 2.0 genes in this pathway |
| PID HDAC CLASSII PATHWAY | 34 | 27 | All SZGR 2.0 genes in this pathway |
| PID AR PATHWAY | 61 | 46 | All SZGR 2.0 genes in this pathway |
| PID HDAC CLASSI PATHWAY | 66 | 50 | All SZGR 2.0 genes in this pathway |
| PID RANBP2 PATHWAY | 11 | 11 | All SZGR 2.0 genes in this pathway |
| PID CMYB PATHWAY | 84 | 61 | All SZGR 2.0 genes in this pathway |
| PID SMAD2 3PATHWAY | 17 | 13 | All SZGR 2.0 genes in this pathway |
| REACTOME MEIOSIS | 116 | 81 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE | 421 | 253 | All SZGR 2.0 genes in this pathway |
| REACTOME CHROMOSOME MAINTENANCE | 122 | 80 | All SZGR 2.0 genes in this pathway |
| REACTOME MEIOTIC SYNAPSIS | 73 | 57 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC3 TARGETS DN | 536 | 332 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY SERUM DEPRIVATION UP | 552 | 347 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN UP | 1142 | 669 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN UP | 612 | 367 | All SZGR 2.0 genes in this pathway |
| MARTIN INTERACT WITH HDAC | 44 | 31 | All SZGR 2.0 genes in this pathway |
| LI AMPLIFIED IN LUNG CANCER | 178 | 108 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA2 PCC NETWORK | 423 | 265 | All SZGR 2.0 genes in this pathway |
| PUJANA CHEK2 PCC NETWORK | 779 | 480 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS DN | 1024 | 594 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 6HR DN | 514 | 330 | All SZGR 2.0 genes in this pathway |
| KAUFFMANN DNA REPAIR GENES | 230 | 137 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 16P13 AMPLICON | 120 | 49 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY MUTATED AND AMPLIFIED IN BREAST CANCER | 94 | 60 | All SZGR 2.0 genes in this pathway |
| FLECHNER PBL KIDNEY TRANSPLANT OK VS DONOR UP | 151 | 100 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| KAAB FAILED HEART ATRIUM DN | 141 | 99 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 48HR DN | 504 | 323 | All SZGR 2.0 genes in this pathway |
| SESTO RESPONSE TO UV C1 | 72 | 45 | All SZGR 2.0 genes in this pathway |
| JIANG HYPOXIA CANCER | 83 | 52 | All SZGR 2.0 genes in this pathway |
| RIZKI TUMOR INVASIVENESS 3D UP | 210 | 124 | All SZGR 2.0 genes in this pathway |
| CHAUHAN RESPONSE TO METHOXYESTRADIOL DN | 102 | 65 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA CHEMOTAXIS DN | 457 | 302 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA HAPTOTAXIS DN | 668 | 419 | All SZGR 2.0 genes in this pathway |
| ROME INSULIN TARGETS IN MUSCLE UP | 442 | 263 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-188 | 29 | 35 | m8 | hsa-miR-188 | CAUCCCUUGCAUGGUGGAGGGU |
| miR-200bc/429 | 256 | 262 | m8 | hsa-miR-200b | UAAUACUGCCUGGUAAUGAUGAC |
| hsa-miR-200c | UAAUACUGCCGGGUAAUGAUGG | ||||
| hsa-miR-429 | UAAUACUGUCUGGUAAAACCGU | ||||
| miR-30-5p | 52 | 59 | 1A,m8 | hsa-miR-30a-5p | UGUAAACAUCCUCGACUGGAAG |
| hsa-miR-30cbrain | UGUAAACAUCCUACACUCUCAGC | ||||
| hsa-miR-30dSZ | UGUAAACAUCCCCGACUGGAAG | ||||
| hsa-miR-30bSZ | UGUAAACAUCCUACACUCAGCU | ||||
| hsa-miR-30e-5p | UGUAAACAUCCUUGACUGGA | ||||
| miR-324-5p | 329 | 336 | 1A,m8 | hsa-miR-324-5p | CGCAUCCCCUAGGGCAUUGGUGU |
| miR-410 | 549 | 555 | 1A | hsa-miR-410 | AAUAUAACACAGAUGGCCUGU |
| miR-485-5p | 453 | 459 | 1A | hsa-miR-485-5p | AGAGGCUGGCCGUGAUGAAUUC |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.