Gene Page: ALDH1A2
Summary ?
| GeneID | 8854 |
| Symbol | ALDH1A2 |
| Synonyms | RALDH(II)|RALDH2|RALDH2-T |
| Description | aldehyde dehydrogenase 1 family member A2 |
| Reference | MIM:603687|HGNC:HGNC:15472|Ensembl:ENSG00000128918|HPRD:04733|Vega:OTTHUMG00000132624 |
| Gene type | protein-coding |
| Map location | 15q21.3 |
| Pascal p-value | 0.823 |
| Sherlock p-value | 0.335 |
| Fetal beta | -0.566 |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenics,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 3 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs6693444 | chr1 | 98690213 | ALDH1A2 | 8854 | 0.08 | trans | ||
| rs16909877 | chr9 | 98216644 | ALDH1A2 | 8854 | 0.12 | trans | ||
| rs9303674 | chr17 | 32458848 | ALDH1A2 | 8854 | 0.16 | trans | ||
| rs12951337 | chr17 | 58154206 | ALDH1A2 | 8854 | 0.09 | trans | ||
| rs17255719 | chrX | 12144766 | ALDH1A2 | 8854 | 0.1 | trans |
Section II. Transcriptome annotation
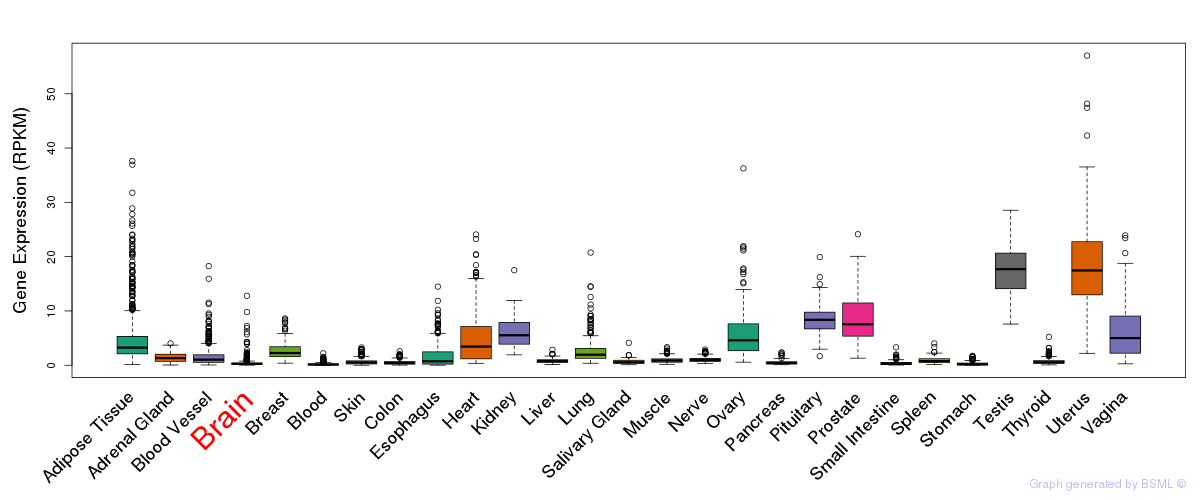
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SCYL1 | 0.90 | 0.87 |
| GRINA | 0.88 | 0.86 |
| ATP6AP1 | 0.88 | 0.88 |
| C3orf39 | 0.87 | 0.85 |
| PNPLA6 | 0.87 | 0.84 |
| ACP2 | 0.86 | 0.87 |
| ATP6V0C | 0.86 | 0.87 |
| AP2A1 | 0.86 | 0.86 |
| CAPNS1 | 0.86 | 0.83 |
| GPI | 0.86 | 0.84 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.21 | -0.62 | -0.44 |
| AF347015.18 | -0.56 | -0.44 |
| AF347015.8 | -0.55 | -0.40 |
| MT-CO2 | -0.54 | -0.39 |
| AF347015.2 | -0.54 | -0.36 |
| MT-ATP8 | -0.53 | -0.42 |
| AF347015.31 | -0.53 | -0.38 |
| MT-CYB | -0.52 | -0.37 |
| NOSTRIN | -0.52 | -0.38 |
| AF347015.26 | -0.51 | -0.35 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0001758 | retinal dehydrogenase activity | IEA | - | |
| GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity | IEA | - | |
| GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity | TAS | 8663198 | |
| GO:0009055 | electron carrier activity | TAS | 8797830 | |
| GO:0016491 | oxidoreductase activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0030182 | neuron differentiation | IEA | neuron (GO term level: 8) | - |
| GO:0030902 | hindbrain development | IEA | Brain (GO term level: 8) | - |
| GO:0030900 | forebrain development | IEA | Brain (GO term level: 8) | - |
| GO:0003007 | heart morphogenesis | IEA | - | |
| GO:0009954 | proximal/distal pattern formation | IEA | - | |
| GO:0009952 | anterior/posterior pattern formation | IEA | - | |
| GO:0008284 | positive regulation of cell proliferation | IEA | - | |
| GO:0009855 | determination of bilateral symmetry | IEA | - | |
| GO:0008152 | metabolic process | IEA | - | |
| GO:0014032 | neural crest cell development | IEA | - | |
| GO:0016331 | morphogenesis of embryonic epithelium | IEA | - | |
| GO:0031016 | pancreas development | IEA | - | |
| GO:0042573 | retinoic acid metabolic process | IEA | - | |
| GO:0048384 | retinoic acid receptor signaling pathway | IEA | - | |
| GO:0043010 | camera-type eye development | IEA | - | |
| GO:0035115 | embryonic forelimb morphogenesis | IEA | - | |
| GO:0055114 | oxidation reduction | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005737 | cytoplasm | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG RETINOL METABOLISM | 64 | 37 | All SZGR 2.0 genes in this pathway |
| GRABARCZYK BCL11B TARGETS DN | 57 | 35 | All SZGR 2.0 genes in this pathway |
| BASAKI YBX1 TARGETS DN | 384 | 230 | All SZGR 2.0 genes in this pathway |
| VECCHI GASTRIC CANCER EARLY DN | 367 | 220 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION HSC UP | 185 | 126 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION MONOCYTE UP | 204 | 140 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION ERYTHROCYTE UP | 157 | 104 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION SUSTAINED IN MONOCYTE UP | 21 | 15 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION SUSTAINDED IN ERYTHROCYTE UP | 44 | 30 | All SZGR 2.0 genes in this pathway |
| DELYS THYROID CANCER DN | 232 | 154 | All SZGR 2.0 genes in this pathway |
| MARKEY RB1 ACUTE LOF DN | 228 | 137 | All SZGR 2.0 genes in this pathway |
| CONCANNON APOPTOSIS BY EPOXOMICIN UP | 239 | 157 | All SZGR 2.0 genes in this pathway |
| MAHADEVAN IMATINIB RESISTANCE DN | 20 | 11 | All SZGR 2.0 genes in this pathway |
| WONG ENDMETRIUM CANCER DN | 82 | 53 | All SZGR 2.0 genes in this pathway |
| EBAUER TARGETS OF PAX3 FOXO1 FUSION UP | 207 | 128 | All SZGR 2.0 genes in this pathway |
| YORDY RECIPROCAL REGULATION BY ETS1 AND SP100 DN | 87 | 48 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA2 PCC NETWORK | 423 | 265 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 48HR UP | 487 | 286 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| YAO HOXA10 TARGETS VIA PROGESTERONE DN | 19 | 12 | All SZGR 2.0 genes in this pathway |
| URS ADIPOCYTE DIFFERENTIATION UP | 74 | 51 | All SZGR 2.0 genes in this pathway |
| KAAB HEART ATRIUM VS VENTRICLE UP | 249 | 170 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| NIELSEN GIST AND SYNOVIAL SARCOMA UP | 20 | 15 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL DN | 216 | 143 | All SZGR 2.0 genes in this pathway |
| ALONSO METASTASIS UP | 198 | 128 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3 | 1069 | 729 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3K4ME2 AND H3K27ME3 | 349 | 234 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN NPC HCP WITH H3K27ME3 | 341 | 243 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 11 | 103 | 68 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS UP | 745 | 475 | All SZGR 2.0 genes in this pathway |
| SCHMIDT POR TARGETS IN LIMB BUD DN | 8 | 7 | All SZGR 2.0 genes in this pathway |
| PLASARI TGFB1 TARGETS 10HR UP | 199 | 143 | All SZGR 2.0 genes in this pathway |
| RAO BOUND BY SALL4 ISOFORM A | 182 | 108 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 2ND EGF PULSE ONLY | 1725 | 838 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-137 | 1744 | 1750 | 1A | hsa-miR-137 | UAUUGCUUAAGAAUACGCGUAG |
| miR-203.1 | 502 | 508 | 1A | hsa-miR-203 | UGAAAUGUUUAGGACCACUAG |
| miR-23 | 1688 | 1695 | 1A,m8 | hsa-miR-23abrain | AUCACAUUGCCAGGGAUUUCC |
| hsa-miR-23bbrain | AUCACAUUGCCAGGGAUUACC | ||||
| miR-323 | 1688 | 1694 | 1A | hsa-miR-323brain | GCACAUUACACGGUCGACCUCU |
| miR-329 | 737 | 743 | m8 | hsa-miR-329brain | AACACACCUGGUUAACCUCUUU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.