Gene Page: LDB1
Summary ?
| GeneID | 8861 |
| Symbol | LDB1 |
| Synonyms | CLIM2|NLI |
| Description | LIM domain binding 1 |
| Reference | MIM:603451|HGNC:HGNC:6532|Ensembl:ENSG00000198728|HPRD:09144|Vega:OTTHUMG00000018950 |
| Gene type | protein-coding |
| Map location | 10q24-q25 |
| Pascal p-value | 8.076E-4 |
| Fetal beta | -0.441 |
| DMG | 1 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg17915961 | 10 | 103881480 | LDB1 | 5.404E-4 | 0.467 | 0.048 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
General gene expression (GTEx)
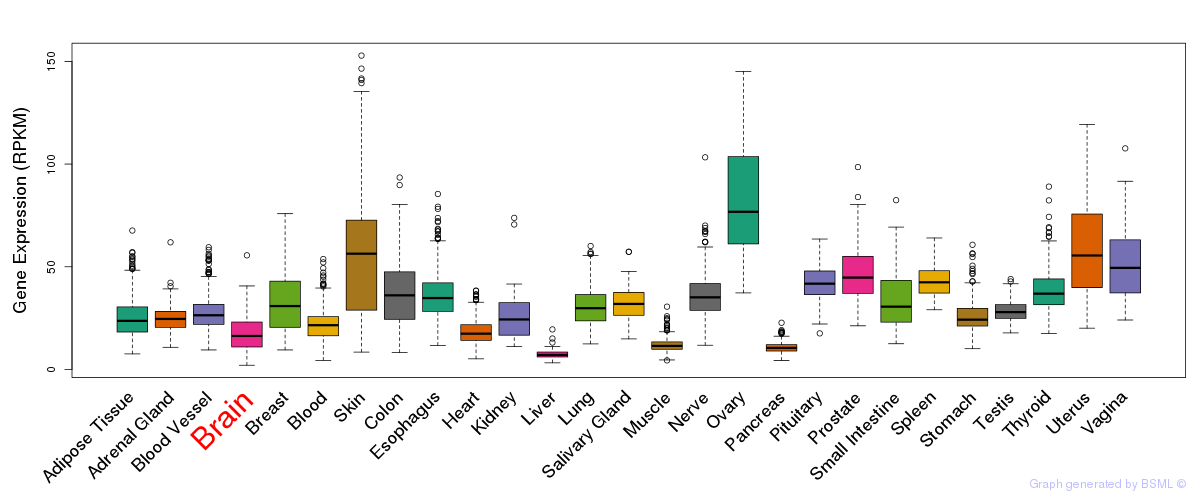
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SMPD4 | 0.95 | 0.95 |
| TSC2 | 0.95 | 0.95 |
| CIZ1 | 0.94 | 0.93 |
| PPRC1 | 0.94 | 0.94 |
| EDC4 | 0.94 | 0.95 |
| PPIL2 | 0.93 | 0.93 |
| DHX34 | 0.93 | 0.92 |
| KIAA0892 | 0.93 | 0.93 |
| MAST2 | 0.93 | 0.92 |
| AC145098.2 | 0.93 | 0.93 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.31 | -0.76 | -0.86 |
| AF347015.27 | -0.74 | -0.84 |
| MT-CO2 | -0.74 | -0.85 |
| AF347015.8 | -0.72 | -0.84 |
| MT-CYB | -0.72 | -0.82 |
| AF347015.33 | -0.72 | -0.80 |
| AF347015.21 | -0.70 | -0.91 |
| AF347015.15 | -0.69 | -0.81 |
| HIGD1B | -0.68 | -0.79 |
| C5orf53 | -0.68 | -0.70 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003714 | transcription corepressor activity | TAS | 9391090 | |
| GO:0030274 | LIM domain binding | IPI | 12792813 | |
| GO:0030274 | LIM domain binding | ISS | - | |
| GO:0042803 | protein homodimerization activity | ISS | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0030182 | neuron differentiation | ISS | neuron (GO term level: 8) | - |
| GO:0007275 | multicellular organismal development | NAS | 12792813 | |
| GO:0045647 | negative regulation of erythrocyte differentiation | ISS | - | |
| GO:0045892 | negative regulation of transcription, DNA-dependent | IDA | 10767331 | |
| GO:0045892 | negative regulation of transcription, DNA-dependent | ISS | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | ISS | - | |
| GO:0043234 | protein complex | ISS | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| BIOCARTA PITX2 PATHWAY | 15 | 15 | All SZGR 2.0 genes in this pathway |
| HUTTMANN B CLL POOR SURVIVAL UP | 276 | 187 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED UP | 633 | 376 | All SZGR 2.0 genes in this pathway |
| PEREZ TP53 TARGETS | 1174 | 695 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN UP | 479 | 299 | All SZGR 2.0 genes in this pathway |
| YAGI AML FAB MARKERS | 191 | 131 | All SZGR 2.0 genes in this pathway |
| LEE CALORIE RESTRICTION NEOCORTEX UP | 83 | 66 | All SZGR 2.0 genes in this pathway |
| MEDINA SMARCA4 TARGETS | 44 | 29 | All SZGR 2.0 genes in this pathway |
| KIM GASTRIC CANCER CHEMOSENSITIVITY | 103 | 64 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| SANSOM APC MYC TARGETS | 217 | 138 | All SZGR 2.0 genes in this pathway |
| SANSOM APC TARGETS REQUIRE MYC | 210 | 123 | All SZGR 2.0 genes in this pathway |
| SANSOM WNT PATHWAY REQUIRE MYC | 58 | 43 | All SZGR 2.0 genes in this pathway |
| IWANAGA CARCINOGENESIS BY KRAS PTEN UP | 181 | 112 | All SZGR 2.0 genes in this pathway |
| SAGIV CD24 TARGETS UP | 23 | 15 | All SZGR 2.0 genes in this pathway |
| SAGIV CD24 TARGETS DN | 46 | 26 | All SZGR 2.0 genes in this pathway |
| CHANG CORE SERUM RESPONSE DN | 209 | 137 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH 11Q23 REARRANGED | 351 | 238 | All SZGR 2.0 genes in this pathway |
| PEDRIOLI MIR31 TARGETS UP | 221 | 120 | All SZGR 2.0 genes in this pathway |
| RAO BOUND BY SALL4 ISOFORM B | 517 | 302 | All SZGR 2.0 genes in this pathway |
| RAO BOUND BY SALL4 | 227 | 149 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-125/351 | 121 | 127 | 1A | hsa-miR-125bbrain | UCCCUGAGACCCUAACUUGUGA |
| hsa-miR-125abrain | UCCCUGAGACCCUUUAACCUGUG | ||||
| miR-365 | 282 | 288 | m8 | hsa-miR-365 | UAAUGCCCCUAAAAAUCCUUAU |
| miR-96 | 151 | 157 | m8 | hsa-miR-96brain | UUUGGCACUAGCACAUUUUUGC |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.